
Section Abstract Introduction Methods Results Discussion Conflict of Interest Acknowledgment References
Review Article
Epigenetic: A new approach to etiology of infertility
pISSN: 0853-1773 • eISSN: 2252-8083
http://dx.doi.org/10.13181/mji.v25i4.1504 Med J Indones. 2016;25:255–62
Received: July 20, 2016
Accepted: December 19, 2016
Author affiliation:
1 Department of Medical Biology, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
2 Biomedical Science, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
Corresponding author:
Silvia W. Lestari
E-mail: finallysilvia@gmail.com
Infertility is a complex disease which could be caused by male and female factors. The etiology from both factors needs further study. There are some approaches to understanding the etiology of infertility, one of them is epigenetic. Epigenetic modifications consist of DNA methylation, histone modifications, and chromatin remodelling. Male and female germinal cells undergo epigenetic modifications dynamically during differentiation into matured sperm and oocyte cells. In a male, the alteration of DNA methylation in spermatogenesis will cause oligo/asthenozoospermia. In addition, the histone methylation, acetylation, or other histone modification may lead sperm lose its ability to fertilize oocyte. Similarly, in a female, the alteration of DNA methylation and histone modification affects oogenesis, created aneuploidy in fertilized oocytes and resulted in embryonic death in the uterus. Alteration of these epigenetic modification patterns will cause infertility, both in male and female.
Keywords
epigenetic alteration, epigenetic modification, female infertility, male infertility
Infertility is defined as the inability to conceive after one year of regular unprotected sexual intercourse by a couple.1 Identifying factors involved in the etiology of infertility is a necessary. There are several approaches to understanding the etiology of infertility, one of which is epigenetics.
Epigenetics is a modified form of chromatin which may alter the structure of chromatin constituent. Changes in chromatin structure will affect the activation or repression of gene expression. Epigenetic changes affect gene expression or phenotype that can be inherited to the next generation without any modification of its deoxyribonucleic acid (DNA) sequences.
Epigenetics regulates a variety of important processes in the body. One of them is fertility which is associated with spermatogenesis or oogenesis. Spermatogenesis is the process of formation, development, and maturation of male germinal cells that occurs in the testis. Oogenesis is the process of formation, development, and oocyte maturation that occurs in the ovary. The newly formed sperms and oocytes undergo epigenetic modifications during the process of differentiation into matured sperms and oocytes. Epigenetics on spermatogenesis and oogenesis is a new issue and there are still many which have not been revealed in the research.
Alteration of these epigenetic patterns can cause infertility, both in male and female. This paper will discuss some of the epigenetic modifications that occur during the process of spermatogenesis and oogenesis as well as some alteration patterns of epigenetic modifications.
Overview of epigenetic mechanism
Epigenetics is defined as the changes that affect gene expression or phenotype and can be inherited to the next generation through meiosis or mitosis without modification in its DNA sequence.2–4 Epigenetics occurs at the level of chromatin, the constituent subunits of nucleosome. The placement of DNA strands that wrapped around histone proteins in chromatin determines whether a gene is transcribed or not. Chromatin is functional in two forms, namely 1) the DNA strand which is compacted tightly during mitosis and meiosis and called heterochromatin. This section is not actively transcribed; and 2) DNA strands that loosely bind to histones and called euchromatin. This section is actively transcribed.5,6
Related to this, epigenetic alteration plays an important role in determining which genes will be expressed in a specific cell. The mechanism of epigenetic modifications in determining the activation and inactivation of genes, occurs through DNA methylation, histone modifications, and chromatin remodeling as described in Figure 1.
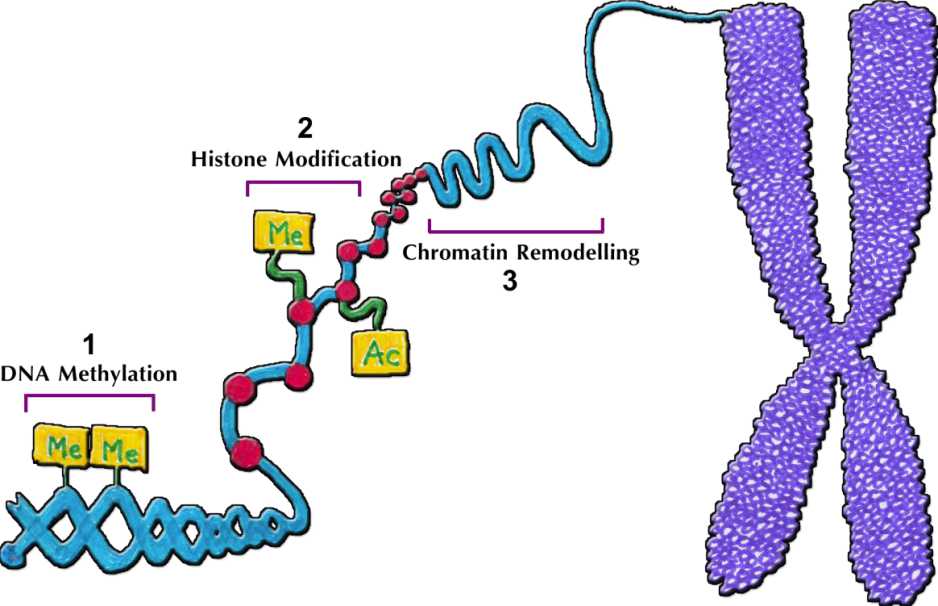
Figure 1. Epigenetic mechanism namely 1) DNA methylation; 2) histone modification; 3) chromatin remodelling
DNA methylation
DNA methylation occurs through the addition of a methyl group of S-adenosylmethionine (SAM) donor to fifth carbon position (C5) major groove of cytosine residues that are in the 5’-G-phosphate-C-3’ (CpG) dinucleotide. CpG dinucleotide consists of cytosine that binds to guanine through phosphodiester bonds and can be found as a group called the CpG island (CGI). CGI associated with gene promoter and regulatory regions in the transcription initiation.7,8
The mechanism of DNA methylation, conducted by the addition of a methyl group to DNA, was catalyzed by the DNA methyltransferase (DNMT) enzymes, whereas DNA demethylation was catalyzed by the DNA demethylation enzymes. In mammals, there are three kinds of DNMT, namely i) DNMT1 which plays role in maintaining methylation, ii) DNMT3a and iii) DNMT3b which is de novo methyltranferases that methylates the genomic DNA during early embryonic development.9 The DNA methylation regulates gene expression by inhibiting the interaction of transcription factors to DNA by compressing the structure of chromatin so transcription factors do not have access to DNA.5,6,10
Histone modification
Histone modification involves nucleosomal histone that has a function in DNA packaging. The visible strands of DNA are wrapped around octamer on two subunits per each core histone (H2A, H2B, H3 and H4) and are connected by connecting histones (H1) to form nucleosomes which become the basic subunit of chromatin. The N-terminal tail of histone consists of amino acid residues which can be affected by methylation, acetylation, phosphorylation, ubiquitylation, and sumoylation.11
Histone methylation is the addition of methyl groups on lysine (K) and arginine (R) residues which is regulated by histone methyltransferase (HMTase). In general, histone methylation is associated with gene silencing, depends on the position of the methylated residue. For example, the lysine 9 on H3 (H3K9) which is methylated, will suppress the gene expression.5,7 On the other hand, the methylation of lysine 4 residue in H3 (H3K4) leads to gene expression.12
Histone acetylation is the transfer of an acetyl group from acetyl-CoA to the cluster ε-NH2 to lysine N-terminal histone residues, catalyzed by the histone acetyltransferase (HAT) enzyme. It activates gene expression and histone deacetylase (HDAC) enzyme that play a role in the deacetylation of histones which will inhibit the gene expression.5,6,11,13,14
Histone phosphorylation is the addition of a phosphate group to the histone proteins. Phosphorylation occurs on serine residues and generally leads to gene activation.15 Histone ubiquitylation the addition of ubiquitin group on the histone proteins. Ubiquitin is a polypeptide consisting of 76 amino acids which can be attached enzymatically to all kinds of proteins. Sumoylation is one of histone modifications with the attachment of small ubiquitin-related modifier protein (SUMO).11 The histone modification by sumoylation may cause gene expression and inhibits other types of histone modification.16
Chromatin remodelling
The third epigenetic mechanism is chromatin remodelling that controls the position of nucleosomes. Chromatin remodelling is regulated by ATP-dependent chromatin remodeling complex, which mediates the access of transcription factors to DNA chromosome as its main function, so it can control gene expression, DNA replication, and DNA repair.5,17
ATP-dependent chromatin remodelling complex requires the energy from ATP hydrolysis to change the position and the structure of the nucleosome, causing the windings of DNA on histones of octamer loosened. As a result, the gene becomes accessible to transcription factors that lead to gene expression or silencing.17
Chromatin remodelling is catalyzed by an ATPase enzyme which is Sucrose Non-Fermentable 2 (SNF2) family. Some members of the SNF2 family like Sth1 subunit of Remodel the Structure of Chromatin (RSC) complex in yeast, Brahma in Drosophila, and Brahma-related gene (BRG)-1 and hBRMin humans also have bromodomain, which can be affected by histone acetylation to encourage chromatin remodeling.17–19
Epigenetic in male infertility
It is estimated that the male factors that cause infertility is about 30%.1 The male factors in infertility could be the abnormality of sperm count (oligozoospermia), the sperm motility (asthenozoospermia), and the sperm morphology (teratozoospermia) or combination, as a result of disruption in spermatogenesis. Epigenetics becomes one of the important roles in the regulation of male fertility, starts from spermatogenesis until embryo development.
DNA methylation and male infertility
De novo DNA methylation occurs in the early stages of spermatogenesis. As explained above, DNA methyltransferase (DNMTs) enzymes regulate de novo DNA methylation, including during spermatogenesis stages. A study showed the highest level of DNMT3a and DNMT3b expression in spermatogonia A.20 The expression of DNMT1 exhibited a similar pattern to DNMT3a and DNMT3b expression.
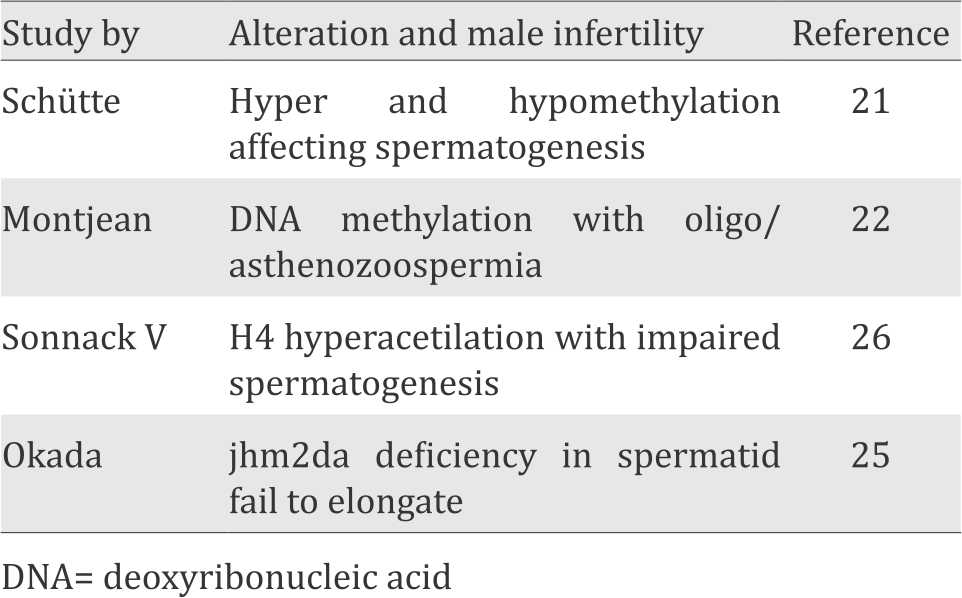
The alteration of the DNA methylation level in the sperm affects male infertility. As in the study Schütte et al21 in men with oligozoospermia, there were several genes that changed in DNA methylation level leading to hypermethylation or hypomethylation which might affect spermatogenesis.
In addition, the research of Montjean et al22 showed that global DNA methylation in sperm was associated with sperm concentration. Oligozoospermic men had global DNA methylation level which were significantly lower compared to normozoospermic men. Furthermore, the global DNA methylation level in sperm was also associated with sperm motility. Men with severe asthenozoospermia had a global DNA methylation level which was significantly lower compared to normal sperm motility.
Histone modification and male infertility
Histone modification that occurs in spermatogenesis could modify the accessibility of DNA to transcription factors. The histone methyltransferase (HMT) and histone demethylase (HDM) enzymes regulate the methylation patterns in histones. Methylation at H3K4 is generally associated with gene expression. In a study conducted by Godmann et al.23 showed the level of H3K4 methylation that had a peak in spermatogonia and diminished in spermatocytes stage. This modification may be required in the protein forming which needed to begin the development phase to become spermatocytes.
H3K9 methylation occurs during meiosis,24 and the process is continued to H3K9 demethylation at the end of meiosis. In his study, Okada et al25 did knockout on Jumonji C (JmjC) domain–containing histone demethylase 2A (jhdm2a) and found that jhdm2a was expressed at round spermatid stage. Furthermore, it was found that jhdm2a deficiency would cause spermatid failure to elongate because of the disruption of chromatin condensation, smaller size of testis, and infertility. Thus, it can be concluded that H3K9 demethylation has an essential role during spermiogenesis process.
During spermatogenesis, H4 hyperacetylation lysine residue that is regulated by the Histone Acetyltransferase (HAT) and Histone Deacetylase (HDAC) enzymes plays an important role in the transition of histone to protamine and allows nucleosomes to experience the demolition at spermatid elongation. The decreased level of H4 hyperacetylation was found in infertile men with impaired spermatogenesis.26 Thus, the ubiquitylation of histones H2A through RNF8-dependent is also an important factor to regulate the demolition of nucleosomes in the post-meiotic stages and in the incorporation of transition protein into chromatin. The ubiquitylation of histone H2A with RNF8 will open conformation of chromatin to become less tight, so as to provide access for the exchange of histone proteins with a protein transition.27 Meanwhile, a study by Lu et al,28 indicated an association between the ubiquitylation of histones H2A and H2B by RNF8 in induces of histone acetylation of H4K16 that facilitated the transition of histone-protamine on spermatid elongation.
Deficiency of RNF8 can cause sperm to become immotile, decrease the number of sperm in the epididymis, make the head of the sperm less condensed, and cause the residual body unseparated completely from the sperm. Furthermore, the results of in vitro fertilization test conducted by Lu et al28 showed that abnormal sperm had no capability to fertilize the normal oocyte.
Sumoylation and male infertility
Another form of epigenetic modifications that occurs during spermatogenesis is sumoylation. Some important proteins in spermatogenesis are known to be SUMO interaction target. Sumoylation has a role as a major factor in the regulation of transcription, response to stress, regulation of enzymatic main pathway, nuclear - cytoplasmic transport, cell cycle control, and other functions in spermatogenesis. Proteins involved in ubiquitylation, DNA repair, and chromatin remodeling experience sumoylation high levels in spermatocytes. In contrast, the cytoskeleton proteins is modified by SUMO in spermatid.29 One of the proteins involved in epigenetic modifications in spermatogenesis is SUMO-1. SUMO-1 expression in testis showed another specific function, namely to help the inactivation of sex chromosomes during meiosis. SUMO-1 is in the chromatin during the zigoten phase when paired homologous chromosomes occurs and during the initiation of sex chromosome condensation.30
The list of epigenetic alterations in male infertility is summarized in Table 1.
Table 1. List of epigenetic alteration in male infertility
Epigenetic in female infertility
Similar to the male factors, it is estimated that the female factors can cause infertility by about 25%. Besides playing a role in male fertility, epigenetics also influence female fertility, i.e. oogenesis. The female infertility factors that are going to be discussed in this paper are ageing oocyte, endometriosis, and polycystic ovarian syndrome (PCOS).31,32 Alterations of epigenetic modification play an essential role in the regulation of gene expression changes in those female factors. Several studies have shown a change in the pattern of DNA methylation and histone modifications in respective genes.
Epigenetic in oogenesis
The oogenesis process in the ovarium is regulated by many factors, i.e. epigenetic mechanism as explain in the Figure 2.
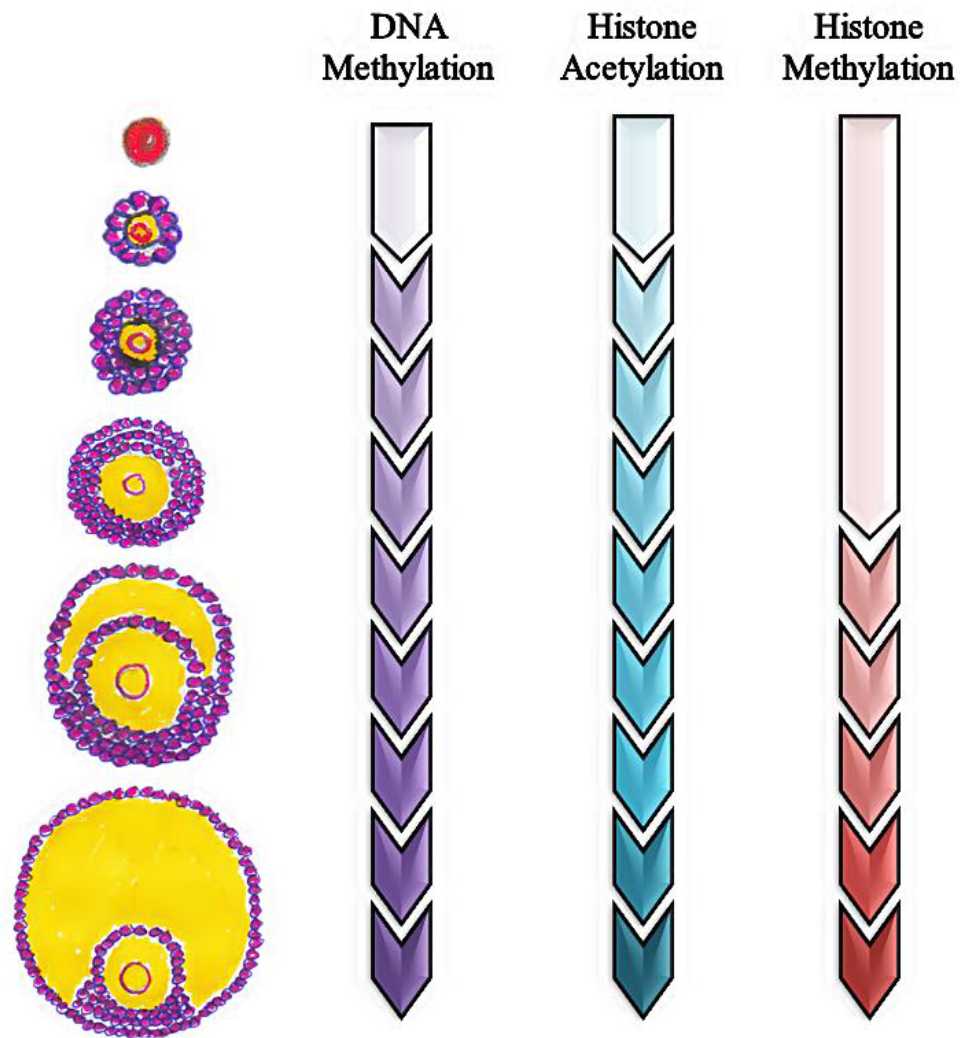
Figure 2. Epigenetic mechanism in the follicle development
The profile of DNA methylation remains constant at the primordial stage to the stage of primary follicles, but it increases progressively until antrum formation. Studies conducted by Kageyama et al34 in the female mice showed the increase of DNA methylation began on day 10 until day 15. Hiura et al35 added that the profile of DNA methylation depended on the size of oocyte, that DNA methylation occured mostly in oocytes with a diameter around 55-60 μm.
In the development of oocytes, besides DNA methylation, there is also histone acetylation. As illustrated, during the early stages of development, histone acetylation has a low level and begins to rise abruptly along with the follicle development.33 Ling Gu et al36 reported the dynamic changes in the acetylation of histone H4 lysine residues at H4K5, H4K8, H4K12, H4K16 positions and histone H3 at H3K9 and H3K14 positions. At the beginning, the level of acetylation in all six residues decreased at the end of the GV stage, indicating that histone deacetylase (HDAC) is required. Furthermore, all of H4K5 and H4K16 have deacetylation on stage MI (metaphase I), followed by reacetylation at the AI stage (anaphase I), then they undergo deacetylation at the MII (metaphase II) of oocyte.
In the oocyte development, when the follicle reached antrum formation, histone H3 dimethylated at K4me2 and K9me2 position or trimethylated at K4me3 position, which is regulated by MLL2 methyltransferase.33 Kageyama et al34 showed that methylation in the H3K4 and H3K9 position was also increased.34 Moreover, H3K4me2, H3K4me3, and H3K9me2 experienced a slight increase in the oocyte development on day 10, and then increased significantly on day 15. The increasing of H3K9 di- and tri-methylation may play a role in the inactivation of the whole genome transcription during oocyte development. In contrast, the H3K4 methylation may be involved in the changes of chromatin configuration, which is not related to transcription.37
Epigenetic and ageing oocytes
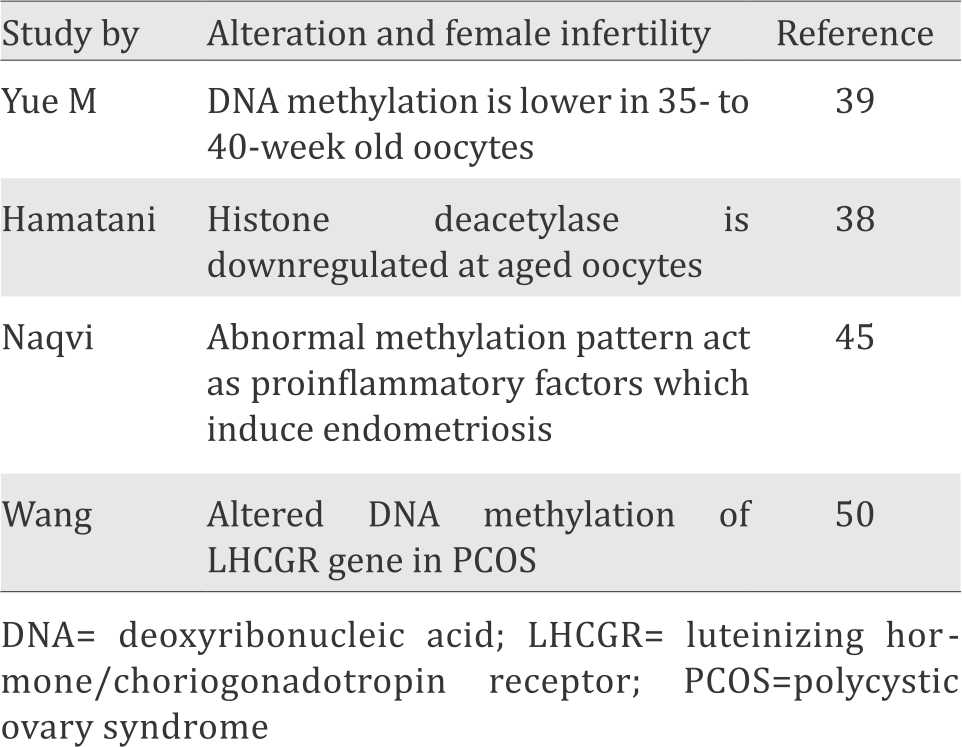
It has been known that the epigenetic modification of oocytes may be affected by advanced maternal age. One of the reasons is associated with the altered expression of DNMTs and histone acetyltransferases. Hamatani et al38 showed the difference of gene expression profile between the 5- to 6-week-old mouse oocytes, related to DNMT1, 3b and 3I activity which maintains the DNA methylation. In addition, the pregnancy rate of older Kunming mice is lower than that of younger ones, which may be associated with abnormal DNA methylation in oocytes.39 Besides the DNA methylation, histone modifications also affect the ageing oocyte. During meiosis, histone is deacetylated globally at the Metaphase I (MI) and Metaphase II (MII) stages by histone deacetylase activity in mammalian oocytes. Histone deacetylase is downregulated at the transcript level in ageing mouse oocytes. In addition, Akiyama et al40 reported that if meiotic histone deacetylation was inhibited, aneuploidy occurred in fertilized mouse oocytes and resulted in embryonic death in the uterus.
Epigenetic and endometriosis
Endometriosis is defined as a disorder in which endometrial tissue grows outside the uterus, which can induce chronic inflammatory reactions.41,42 Epigenetics plays a role in regulating the expression of endometrial genes during the menstrual cycle. The alteration in the epigenetic modification is a mechanism that may occur in endometriosis-induced changes in gene expression in endometriosis, including genes that play a role in the regulation of transcription, proliferation, inflammation, apoptosis, and cell cycle associated with implantation failure.43 In women with eutopic endometriosis, an increased of DNMT3A expression was found, which might play a role in hypermethylation in some conditions of endometriosis.44
Naqvi et al45 found the alteration of DNA methylation in some genes associated with endometriosis. The abnormalities of the methylation pattern of genes that act as proinflammatory factors will cause a change in the signal of the immune response, which induced chronic inflammation in endometriosis. Moreover, the expression of DNA methyltransferase enzyme that plays a role in repairing DNA methylation patterns, has been suppressed due to hypermethylation of its encoding gene, namely MGMT. It will create the possibility of DNA methylation irregularities of candidate genes in endometriosis. Meanwhile, other irregularities of gene expression resulting from changes in DNA methylation become hypermethylated which occured in the progesterone-B, E-cadherin and HOXA10 gene receptor. In addition, estrogen-β receptor, steroidogenic-1factor and aromatase also underwent hypomethylation.46
Inhibition of the E-cadherin expression due to hypermethylation causes disruption of cell adhesion, thus inducing invasion and metastasis of endometrial cells. HOXA10 genes in humans is expressed in the endometrium that is regulated by estrogen and progesterone. HOXA10 plays role in the development of the endometrium to prepare for implantation. Hypermethylation on HOXA10 causes a defect in the endometrium function, hence implantation failure in women with endometriosis. Meanwhile, aromatase is an enzyme that catalyzes the conversion of androgens to estrogens. Hypomethylation on aromatase gene leads to the increase of aromatase expression and of estrogen production that induces proliferation of endometrium cell.46,47
Meanwhile, deviations of other epigenetic modification, such as acetylation of histone modifications are also found in endometriosis. Histone acetylation is generally associated with the activation of a gene expression. The analysis of the chromatin immunoprecipitation (ChIP) conducted by Monteiro et al48 showed the hypoacetylation on the H3/H4 at the promoter regions of candidate genes was down regulated in endometriosis, namely HOXA10, ESR1, CDH1 and p21.48 On the other hand, the steroidogenic factor 1 promoter gene underwent increased acetylation of H3 and H4 which correlated with the high expression of the gene.
Epigenetic and polycystic ovarian syndrome (PCOS)
Polycystic ovarian syndrome (PCOS) is a set of symptoms resulting from the hormonal imbalance in women. It is characterized by the following three conditions: 1) the absence of ovulation, which leads to irregular menstrual periods or amenorrhea, 2) high androgen, and 3) the presence of cysts on one or both ovaries. PCOS is a major cause of infertility as a result of the absence of ovulation.49
The other epigenetic alteration found in PCOS, is a deviation of DNA methylation pattern in multiple genes. Wang et al.50 found an altered level of DNA methylation luteinizing hormone (LH)/choriogonadotropin receptor (LHCGR) gene in patients with PCOS had hypomethylation on its multiple CpG sites. LHCGR is identified as a susceptibility gene for PCOS. LHCGR interaction with its ligands, namely LH, has an important role in the process of ovulation in mammals. Meanwhile, LH induces theca cells in ovarian to secrete androgen precursor.51
Therefore, LHCGR can be considered as a candidate gene of PCOS. Hypomethylation on LHCGR genes causes the expression of LH receptors to increase, which leads to increase of sensitivity to LH.
The list of epigenetic alteration in female infertility is summarized in Table 2.
Table 2. List of epigenetic alteration in female infertility
In conclusion, epigenetic modifications play an important role as regulators in the activation and inactivation of gene expression. Epigenetic modifications during spermatogenesis occur with various dynamic intensities. This review shows that epigenetic modifications play specific roles at certain stages in the spermatogenesis process, from DNA methylation and histone modification to sumoylation. Epigenetic modifications during oogenesis also affect the oocyte quality as well as in the other etiology of female infertility, i.e. endometriosis and PCOS. Alteration in the epigenetic modification patterns is associated with disturbances in spermatogenesis and oogenesis which might lead to infertility.
Conflicts of Interest
The authors affirm no conflict of interest in this study.
Acknowledgment
Authors would like to express gratitude to Indonesian Reproductive Medicine Research and Training Center (Ina-Repromed) for supporting this review.
REFERENCES
- Stanford JB, Parnell TA, Boyle PC. Outcomes from treatment of infertility with natural procreative technology in an Irish general practice. J Am Board Fam Med. 2008;21(5):375–84.
- Carrell DT. Epigenetics of the male gamete. Fertil Steril. 2012;97(2):267–74.
- Güneş S, Kulaç T. The role of epigenetics in spermatogenesis. Turk J Urol. 2013;39(3):181–7.
- Zamudio NM, Chong S, O’Bryan MK. Epigenetic regulation in male germ cells. Reproduction. 2008;136(2):131–46.
- Armstrong L. Epigenetics. New York: Garland Science; 2014.
- Tollefsbol TO. Handbook of epigenetics: the new molecular and medical genetics. In: Tollefsbol TO, editor. Epigenetics: the new science of genetics. London: Academic Press; 2010. p. 1–6.
- Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839(12):1362–72.
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & Dev. 2011;25(10):1010–22.
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19(3):219–20.
- Oh JH, Gertych A, Tajbakhsh J. Nuclear DNA methylation and chromatin condensation phenotypes are distinct between normally proliferating/aging, rapidly growing/immortal, and senescent cells. Oncotarget. 2013;4(3):474–93.
- Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727(3):62–71.
- Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, et al. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121(7):2641–50.
- Peserico A, Simone C. Physical and functional HAT/ HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011(371832):1–10
- Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17(4):430–7
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–8.
- Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100(23):13225–30.
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatinremodeling enzymes. Cell. 2013;154(3):490–503.
- Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Bio. 2002;3(6):422–9.
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25(12):1192–200.
- La Salle S, Trasler JM. Dynamic expression of DNMT3a and DNMT3b isoforms during male germ cell development in the mouse. Dev Biol. 2006;296(1):71–82.
- Schütte B, El Hajj N, Kuhtz J, Nanda I, Gromoll J, Hahn T, et al. Broad DNA methylation changes of spermatogenesis, inflammation and immune response-related genes in a subgroup of sperm samples for assisted reproduction. Andrology. 2013;1(6):822–9.
- Montjean D, Zini A, Ravel C, Belloc S, Dalleac A, Copin H, et al. Sperm global DNA methylation level: association with semen parameters and genome integrity. Andrology. 2015;3(2):235–40.
- Godmann M, Auger V, Ferraroni-Aguiar V, Di Sauro A, Sette C, Behr R, et al. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod. 2007;77(5):754–64.
- Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26(14):3346–59.
- Okada Y, Tateishi K, Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J Androl. 2010;31(1):75–8.
- Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34(6):384–90.
- Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012;26(24):2737–48.
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18(3):371–84.
- Xiao Y, Pollack D, Andrusier M, Levy A, Callaway M, Nieves E, et al. Identification of cell-specific targets of sumoylation during mouse spermatogenesis. Reproduction. 2016;151(2):149–66.
- Vigodner M, Morris PL. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev Biol. 2005;282(2):480–92.
- Derman S, Seifer DB. Evaluation of female infertility. In: Alvero R, editor. Reproductive endocrinology and infertility. Philadelphia: Mosby; 2007. p. 155–68.
- Simpson J. Molecular approach to common causes of female infertility. Best Pract Res Clin Obstet Gynaecol. 2002;16(5):685–702.
- Zuccotti M, Merico V, Redi CA, Garagna S. An epigenomic biography of the mammalian oocyte. In: Coticchio G, Albertini DF, Santis DL, editor. Oogenesis. London: Springer; 2013. p. 141–50.
- Kageyama S, Liu H, Kaneko N, Ooga M, Nagata M, Aoki F. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction. 2007;133(1):85–94.
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11(4):353–61.
- Gu L, Wang Q, Sun Q-Y. Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle. 2010;9(10):1942–50.
- Pan Z, Zhang J, Li Q, Li Y, Shi F, Xie Z, et al. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J Genet Genomics. 2012;39(3):111–23.
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6(1):117–31.
- Yue MX, Fu XW, Zhou GB, Hou YP, DU M, Wang L, et al. Abnormal DNA methylation in oocytes could be associated with a decrease in reproductive potential in old mice. J Assist Reprod Genet. 2012;29(7):643–50.
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A. 2006;103(19):7339–44.
- Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30(3):266–77.
- Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7(10):1098–108.
- Sawicka A, Seiser C. Sensing core histone phosphorylation - a matter of perfect timing. Biochim Biophys Acta. 2014;1839(8):711–8.
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281(29):20643–9.
- Naqvi H, Ilagan Y, Krikun G, Taylor HS. Altered genomewide methylation in endometriosis. Reprod Sci. 2014;21(10):1237–43.
- Nasu K, Kawano Y, Tsukamoto Y, Takano M, Takai N, Li H, et al. Aberrant DNA methylation status of endometriosis: epigenetics as the pathogenesis, biomarker and therapeutic target. J Obstet Gynaecol Res. 2011;37(7):683–95.
- Zanatta A, Rocha AM, Carvalho FM, Pereira RM, Taylor HS, Motta EL, et al. The role of the Hoxa10/ HOXA10 gene in the etiology of endometriosis and its related infertility: a review. J Assist Reprod Genet. 2010;27(12):701–10.
- Monteiro JB, Colón-Diaz M, Garcia M, Gutierrez S, Colón M, Seto E, et al. Endometriosis is characterized by a distinct pattern of histone 3 and histone 4 lysine modifications. Reprod Sci. 2014;21(3):305–18.
- Ndefo UA, Eaton A, Green MR. Polycystic ovary syndrome: a review of treatment options with a focus on pharmacological approaches. PT. 2013;38(6):336.
- Wang P, Zhao H, Li T, Zhang W, Wu K, Li M, et al. Hypomethylation of the LH/choriogonadotropin receptor promoter region is a potential mechanism underlying susceptibility to polycystic ovary syndrome. Endocrinology. 2014;155(4):1445–52.
- Young JM, McNeilly AS. Theca: the forgotten cell of the ovarian follicle. Reproduction. 2010;140(4):489–504.
Copyright @ 2016 Authors. This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original author and source are properly cited.
mji.ui.ac.id