
Section Abstract Introduction Methods Results Discussion Conflict Of Interest Acknowledgment References
Basic Medical Research
Knock-out transmembrane prostate androgen-induced protein gene suppressed triplenegative breast cancer cell proliferation
pISSN: 0853-1773 • eISSN: 2252-8083
http://dx.doi.org/10.13181/mji.v26i3.1823 Med J Indones. 2017;26:178–82
Received: February 7, 2017
Accepted: September 6, 2017
Author affiliation:
1 Doctoral Program in Biomedicine, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
2 Medical Science Master Program, Graduate School of Comprehensive Human Science, University of Tsukuba, Japan
3 Departement of Experimental Pathology, Faculty of Medicine, University of Tsukuba, Japan
4 Departement of Pharmacology and Therapeutics, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia
Corresponding author:
Melva Louisa
E-mail: melva.louisa@gmail.com
Background
Triple negative breast cancer (TNBC) tends to grow more rapidly and has poorer prognosis compared to others. High expression of transmembrane prostate androgen-induced protein (TMEPAI) correlates with poor prognosis in TNBC patients. However, the mechanistic role of TMEPAI in tumorigenic remains unknown. This study aimed to knock-out TMEPAI in TNBC cell line to determine its function further in cells proliferation.
Methods
CRISPR-Cas9 has been used previously to knockout TMEPAI in Hs857T TNBC cell line. Hs587T TNBC parental cell line (wild-type/WT) and TMEPAI knock out Hs 586T cell lines were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin and amphotericin B. Both cell lines were seeded in 24-well plates and counted every two days, then proliferation rates were plotted. Afterwards, total RNA were isolated from the cells and Ki-67, and TGF-β mRNA expression levels as proliferation markers were determined.
Results
Cell proliferation rates as displayed in growth curve plots showed that WT-TMEPAI cell line grew more rapidly than KO-TMEPAI. In accordance, mRNA expression levels of Ki-67 and TGF-β were significantly decreased in KO-TMEPAI cell line.
Conclusion
Knock-out of TMEPAI attenuates cell proliferation in TNBC.
Keywords
proliferation, TGF-β, TMEPAI, triple negative breast cancer
Triple negative breast cancer (TNBC) presents in a small number of patients of 15–25% out of the total breast cancer patients.1,2 However, this type of breast cancer is related to shorter survival, poorer prognosis, and metastasize more frequently in lungs and brains compared to other breast cancer subtypes. 1–3 Until now, there is no available targeted therapy indicated for TNBC due to lack expressions of estrogen, progesterone, and human epidermal receptors. The current modality of treatment is chemotherapy such as doxorubicin and paclitaxel, with no specific targets.3 Unfortunately, this type of breast cancer is more prone to develop resistance, either intrinsic or acquired to cytotoxic agents.4 Therefore, a deep and thorough understanding for the pathogenesis as well as chemo-resistance mechanism is urgent in order to determine a novel drug targets for TNBC patients.
Transmembrane prostate androgen-induced protein (TMEPAI) is remarkable in TNBC. It found highly expressed in 68,8% TNBC patients.2 Moreover, high expression of TMEPAI correlates to poorer prognosis and shorter relapse-free survival.5 The importance of TMEPAI in TNBC started when Singha et al suggested it as a responsible converter for TGF-β from tumor suppressor into promotor.6 Further study by Singha et al2 discussed about the mechanism of TMEPAI in TNBC. TMEPAI inhibits Smaddependent pathways of TGF-β and instantly promotes non-Smad pathways via PI3K/Akt signaling. Previously, Watanabe at al proved how TMEPAI works in Smad-dependent pathways to inhibit TGF-β signaling. TMEPAI disrupts interaction between R-Smad and SARA through Smad interaction motif (SIM). Afterwards, TMEPAI traps phosphorylated Smad2 and Smad3 by preventing their translocation into the nucleus. Consequently, transcription and translation of TGF-β target genes will be stopped.7
In addition, TMEPAI promotes degradation of PTEN, thus promotes PI3K/Akt pathways to be activated. It resulted in an enhancement of cell growth.2 Furthermore, TMEPAI has been proven to increase tumorigenic properties in lung cancer.8 However, the mechanism of how TMEPAI enhances tumorigenic activities in cancer remains unknown. Hence, we aimed to investigate the role TMEPAI in TNBC to determine its function further in cell proliferation, using our previous knock out TMEPAI TNBC.
METHODS
Cell culture and cell proliferation rate
Triple negative breast cancer cell line, Hs578T was obtained from the American Type Culture Collection (ATCC). Hs578T with TMEPAI knockout (KO) cells was developed from HS578T wild-type (WT) previously using CRISPR-Cas9 system.9 Both of Hs578T, WT and KO, were cultured in Dulbecco’s modified essential medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gibco), 10 μg/mL insulin, and 100 units/mL of penicillin G and 0.1 mg/mL of streptomycin sulfate (Wako). Cells were maintained in 5% CO2 incubator at 37°C. Both cell lines were seeded in same number in 24-well plates and counted every two days using hemocytometer in 0.4% trypan blue cell suspension up to 8 days. Area under the curve (AUC) of cell proliferation was calculated by connecting the dots which represents number of cells in different time points. AUC were calculated automatically using GraphPad Prism v. 7.0 software (GraphPad Prism, USA). Doubling time for each cell line were also calculated using online doubling time calculator.10 All of the experiments were conducted three times and were duplicated.
RT-PCR
We isolated total RNA using Total RNA Mini Kit (Geneaid) for cultured cells, and afterwards, converted total RNA into cDNA using Transcriptor First Strand cDNA Synthesis Kit (Roche). RTPCR were performed using FastStart Essential DNA Green Master (Roche). All primers were purchased from the Integrated DNA Technologies, Singapore. Primer sequences all of the mRNA evaluated were as below: Ki-67 Fwd: 5’-TCCTTTGGTGGGCACCTAAGACCTG-3’; Ki-67 Rev: 5’-TGATGGTTGAGGTCGTTCCTTGATG-3’; TGF-β Fwd: 5’-TGAACCGGCTTTTCCTGCTTCTCATG-3’; TGF-β Rev: 5’-GCGGAAGTCAATGTACAGCTGCCGC-3’. β-actin was used as a house-keeping gene with primer sequences: β-actin Fwd: 5’-GCTGGAAGGTGGACAGCGA-3’ and β-actin Rev: 5’-GGCATCGTGATGGACTCCG-3’.
Statistical analysis
Data were shown as mean ± standard deviation (SD). Statistical analysis was performed with independent t-test comparing area under the curve (AUC) for proliferation rate, mRNA expressions of Ki-67, and TGF-β of knockout cell line versus wild-type. Differences were considered statistically significant at p<0.05. All the statistical analysis and graphs presented were calculated and drawn using GraphPad Prism v 7.0 software (GraphPad Prism, USA).
RESULTS
Decreased cell proliferation rate in KOTMEPAI versus WT-TMEPAI TNBC
KO-TMEPAI (Figure 1A) had lower cell confluency compared to WT-TMEPAI Hs587T cells (Figure 1B). In accordance, cell proliferation rate of KOTMEPAI was slower than those of WT-TMEPAI Hs587T cells (Figure 1C), along with lower area under curve (Figure 1D). Doubling time for KOTMEPAI was longer than WT-TMEPAI cells as shown in (Figure 1E).
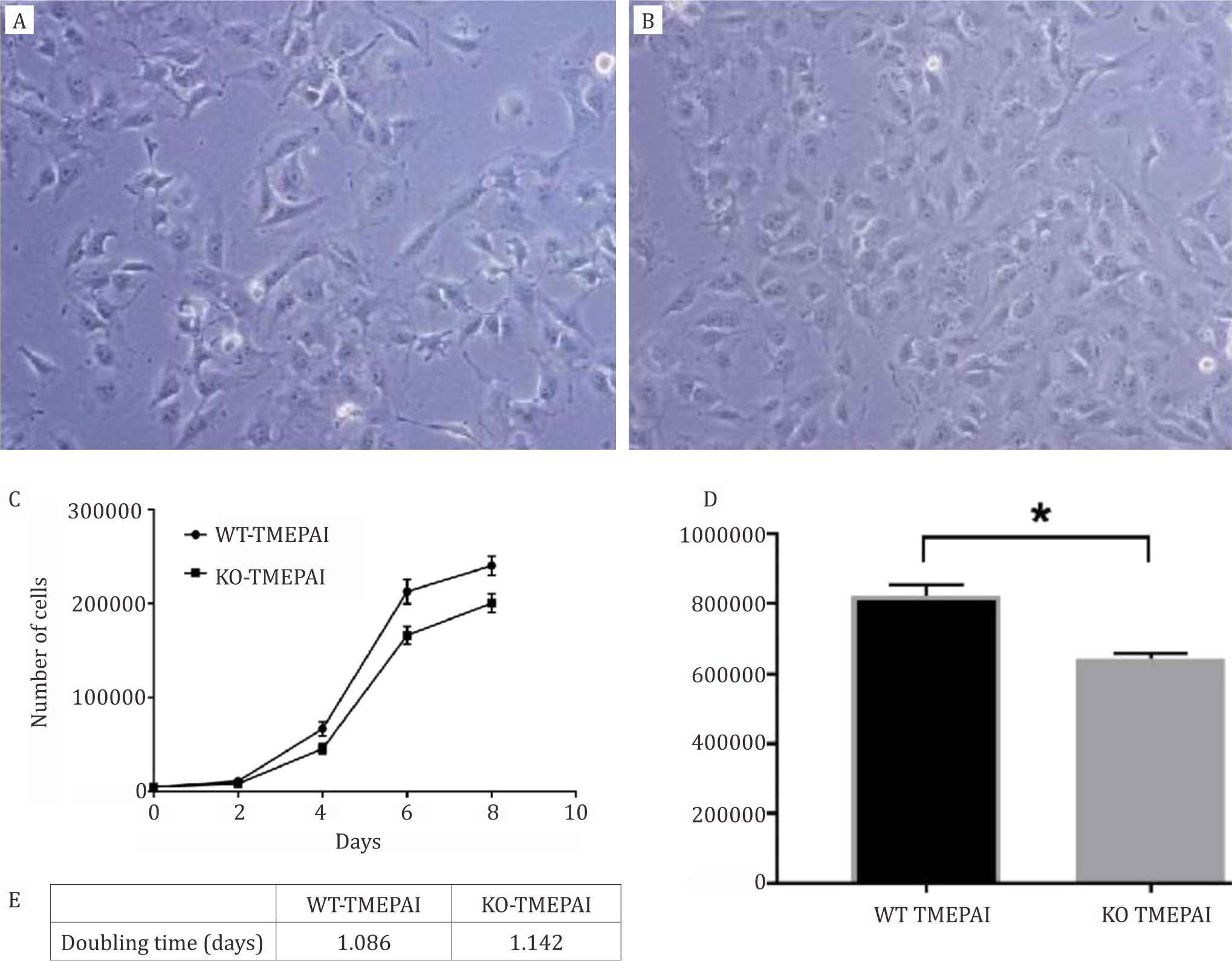
Figure 1. WT-TMEPAI cells grew more rapidly than KO-TMEPAI Hs587T cells. A) TNBC cell line, Hs578T, with wild-type TMEPAI (WT-TMEPAI). B) TNBC cell line, Hs578T, with knock-out TMEPAI (KO-TMEPAI). Cells were taken picture under inverted microscope with 10x magnification. C) Cell proliferation rates of WT-TMEPAI and KO-TMEPAI Hs587T cells were determined using cell growth curve plot up to 8 days. D) Area under curve of WT-TMEPAI and KO-TMEPAI cells proliferation rate. E). Doubling time for KO-TMEPAI was longer compared with WT-TMEPAI Hs587T cells. All experiments were conducted three times in duplicate. WT: wild-type. KO: knock-out. *) p<0.05 after independent t-test comparing wild-type versus knock-out
Proliferation marker and TGF-β were attenuated in the knock-out TMEPAI Hs587T cells
Figure 2A and 2B show alterations of mRNA expressions of Ki-67 and TGF-β. Proliferation marker, Ki-67 decreased remarkably in KOTMEPAI Hs587T cells as well as TGF-β.
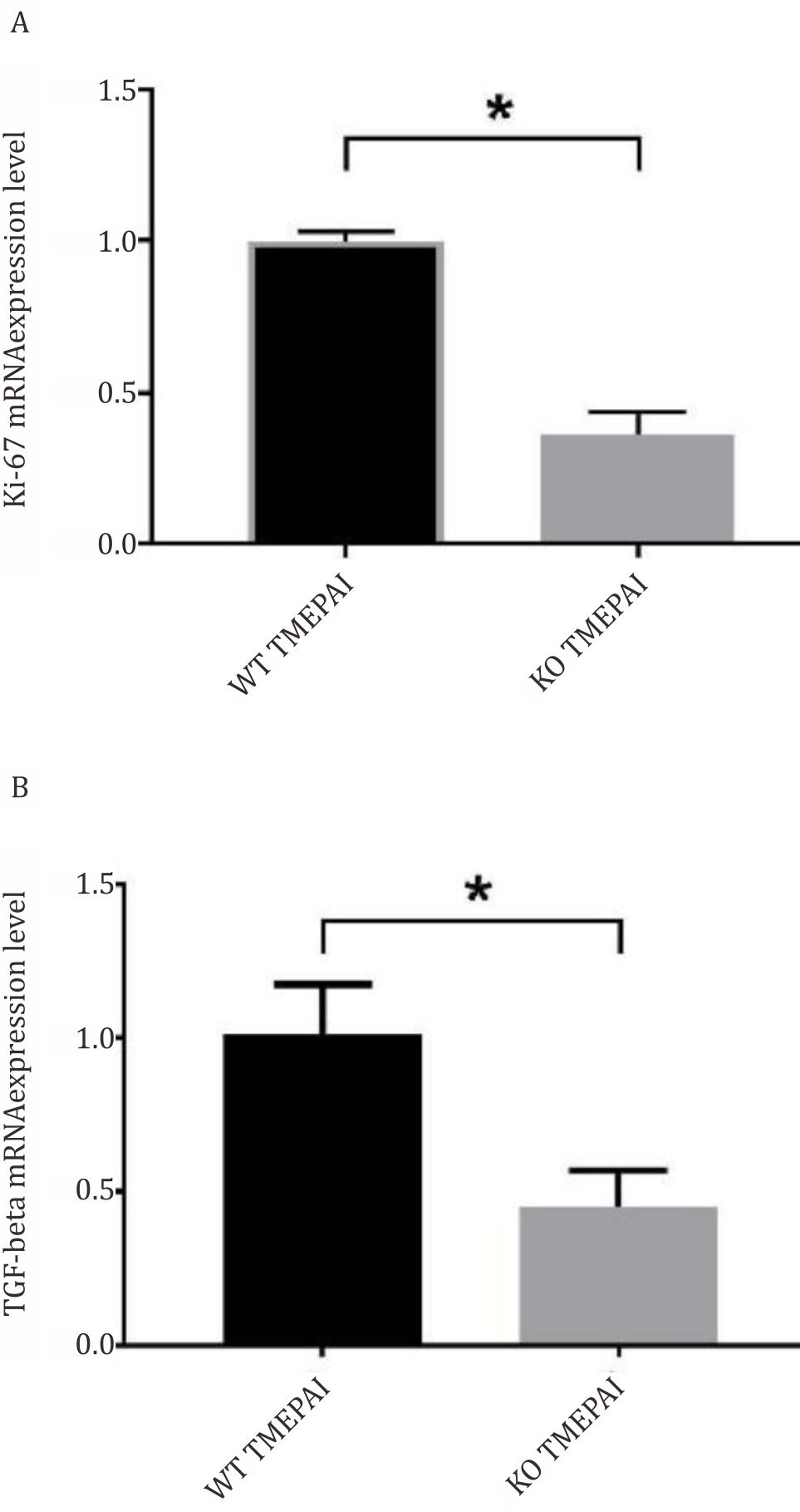
Figure 2. Decreased mRNA expressions of A) Ki-67 (normalized to β-actin as housekeeping gene) and B) TGF-β (normalized to β-actin as housekeeping gene) in KO-TMEPAI as compared to WT-TMEPAI. All experiments were performed three times in duplicate. WT: wild-type. KO: knock-out. *) p<0,05 after independent t-test comparing wild-type versus knock out
DISCUSSION
It is noteworthy that currently there is no targeted therapy registered for triple negative breast cancer (TNBC) worldwide. TNBC is counted due to its more aggressive behavior, and thus, resulted in poorer prognosis as compared to other types of breast cancer. Moreover, TNBC frequently occurs in young woman.3 Many researchers aimed to elucidate the signaling pathways which involved in TNBC as a target for novel therapy.
TMEPAI has been suggested to enhance tumorigenic activity in breast cancers. Previously, there were two reports from Singha2,6 which used knock-down TMEPAI TNBC cells to elucidate the role of TMEPAI in oncogenic and tumorigenic activities of TNBC. However, their model, that used shRNA, could not fully omitted TMEPAI in TNBC cells. Therefore, in this study we aimed to determine cell proliferation alteration in a fully knock-out TMEPAI TNBC cells.
We established knock-out TMEPAI cells in previous study using CRISPR-Cas9 systems in Hs578T wild-type, one of triple negative cell line.9 In this study, we showed that KO-TMEPAI cells had longer doubling time compared to WT-TMEPAI cells. KO-TMEPAI cells also showed a slower proliferation rate than that of wild-type TMEPAI cell line. This strongly indicated that TMEPAI had an important role in cell proliferations of triple negative breast cancer. In accordance, previous study by Singha et al2 reported that knockdown TMEPAI attenuated tumorigenic activity. TMEPAI was reported by Azami et al. as a protumoriogenic factor in lung cancer induced by TGF-β signaling. It also dominantly depended on activated autocrine TGF-β signaling.11
Furthermore, we also evaluated mRNA expression levels of TGF-β. It was previously known that TGF-β induced TMEPAI expression in genomic transcript, as well as proteomic levels.2,6-8 Our study differed with previous study which were conducted by Singha et al2,6 and Ngunyen et al8 Those study used TGF-β induction in cell medium. In our study, we did not add TGF-β exogenously. This condition might be due to the basal conditions that corresponded to autocrine signaling of TGF-β in KO and WT-TMEPAI TNBC cells. Our results in KO-TMEPAI cells confirmed that mRNA expression levels of TGF-β was lower than that of wild-type cells. This indicated that the loss of TMEPAI resulted in decreased proliferation rates, followed by decreased mRNA expressions of TGF-β. The expression of TMEPAI was controlled by TGF-β constituvely. However, TMEPAI suppressed TGF-β signaling by binding to R-Smad competitively with SARA (Smad anchor for receptor activation), to refrain R-Smad binding and inhibit activation of TGF-β receptor kinase.7 It is remain unclear how TMEPAI affects cell proliferation.
In this study, the decrease of T`GF-β mRNA expression levels was still inadequate to describe the activation of TGF-β that was related to cell proliferation signaling. Further studies on TMEPAI downstream targets are still needed. On the other hand, xenograft models from KO-TMEPAI triple negative breast cancer cells should be generated in order to determine the role of TMEPAI in TNBC.
Previous study by Singha et al2 reported that tumor volumes and weights of triple negative xenografts models were less in knock-down TMEPAI group compared to the wild-type group after 28 days. The result was followed by the decrease in proliferation marker, Ki67. In accordance with that finding, our KO-TMEPAI TNBC also showed a decrease in Ki-67 mRNA expressions compared to wild-type cells. Cell proliferation rate and confluency supported that finding. As indicated before, TMEPAI has pivotal role to alter cancer proliferation in triple negative breast cancer.
Therefore, we concluded that TMEPAI attenuates cell proliferation in TNBC cells. In support our finding, Singha et al2, Watanabe et al7 and Ngunyen et al8 already suggested role of TMEPAI in TGF-β signaling pathway through Smad-dependent and PI3K/Akt pathways. Although we have not investigated in xenograft models and determine downstream of TGF-β signaling due to the loss of TMEPAI, we suggest that TMEPAI has a potency to affect cancer progression in triple negative breast cancer, so that it potentially becomes a novel targeted therapy.
Conflicts of Interest
Melva Louisa and Rianto Setiabudy are editorial board members but were not involved in the review or decision for the article.
Acknowledgment
This work was supported by a grant from PITTA (Publikasi Internasional Terindeks untuk Tugas Akhir Mahasiswa) 2016 from Research Institution of Universitas Indonesia (Direktorat Riset dan Pengabdian Kepada Masyarakat, DRPM, Universitas Indonesia).
REFERENCES
- Ng CH, Pathy NB, Taib NA, Teh YC, Mun KS, Amiruddin A, et al. Comparison of breast cancer in Indonesia and Malaysia-A Clinico-Pathological study between Dharmais Cancer Center Jakarta and University Malaya Medical Center, Kuala Lumpur. Asian Pacific J Cancer Prev. 2011;12:2943–6.
- Singha PK, Pandeswara S, Geng H, Lan R, Venkatachalam MA, Saikumar P. TGF-β induced TMEPAI/PMEPA1 inhibits canonical Smad signaling through R-Smad Sequestration and promotes non-canonical PI3K/Akt signaling by reducing PTEN in triple negative breast cancer. Genes&Cancer. 2014;5(9-10):320–36.
- Foulkes WD, Smith IE, Reis-Fielho JS. Triple negative breast cancer. N Eng J Med. 2010;363:1938–48.
- O’Reilly EA, Gubbins L, Sharma S, Tully R, Guang MHZ, Weiner-Gorzel K, et al. The fate of chemoresistance in triple negative breast cancer (TNBC). BBA Clinical 3. 2015;3:257–75.
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010 Oct;123(3):725–31.
- Singha PK, Yeh IT, Venkatachalam MA, Saikumar P. TGF-β-Inducible gene TMEPAI converts TGF- β from a tumor suppressor to a tumor promoter in breast cancer. Cancer Res. 2009;70:6377–83.
- Watanabe Y, Itoh S, Goto T, Ohnishi E, Inamitsu M, Itoh F, et al. TMEPAI, a transmembrane TGF-β-inducible protein, sequesters Smad proteins from active participation in TGF-β signaling. Mol Cell. 2010;37:123–34.
- Ngunyen TTV, Watanabe Y, Shiba A, Noguchi M, Itoh S, Kato M. TMEPAI⁄PMEPA1 enhances tumorigenic activities in lung cancer cells. Cancer Sci. 2014;105(3):334–41.
- Wardhani BWK, Puteri MU, Watanabe Y, Louisa M, Setiabudy R, et al. TMEPAI genome editing in triple negative breast cancer cells. Med J Indones. 2016;26:14–8.
- Roth V. 2006 Doubling Time Computing [Internet]. [cited 2016]. Available from: http://www.doublingtime. com/compute.php
- Azami S, Vo Nguyen T, Watanabe Y, Kato M. Cooperative induction of transmembrane prostate androgeninduced protein TMEPAI/PMEPA1 by transforming growth factor-β and epidermal growth factor signaling. Biochem Biophy Res Commun. 2015;456:580–5.
Copyright @ 2017 Authors. This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original author and source are properly cited.
mji.ui.ac.id