
Section Abstract Introduction Methods Results Discussion Conflict Of Interest Acknowledgment References
Basic Medical Research
The production of SPusp45-MSP-119 gene construct and its recombinant protein in Lactococcus lactis to be used as a malaria vaccine
pISSN: 0853-1773 • eISSN: 2252-8083
https://doi.org/10.13181/mji.v26i4.2162 Med J Indones. 2017;26:261–9
Received: July 19, 2017
Accepted: December 29, 2017
Author affiliation:
1 Research Center for Biotechnology, Indonesian Institute of Science (LIPI), Bogor, Indonesia
2 School of Biotechnology, Bogor Agricultural University, Bogor, Indonesia
3 Research Center for Bioresources and Biotechnology, Bogor Agricultural University, Bogor, Indonesia
Corresponding author:
Apon Z. Mustopa
E-mail: azmustopa@yahoo.com
Background
Merozoite surface protein 1 (MSP-1) is a major protein used by the Plasmodium during red blood cells invasion in malaria. MSP-119, one of MSP-1 is highly conserved, and it is a potential malaria vaccine candidate because the monoclonal antibodies are capable blocking erythrocyte invasion in vitro. The aim of this study was to produce MSP-119 gene construct and the recombinant protein in Lactococcus lactis.
Methods
Usp45-MSP-1 19, derived from codon optimization and the synthetic gene, was inserted into the pMAT cloning vector. A vector expressing MSP-119 included usp45 has been constructed by the manipulation of recombinant DNA using restriction enzymes. The MSP-119 protein was expressed to 45% ammonium sulfate precipitation and purified using Sephadex-G50 gel filtration chromatography. The expressed protein was characterized by SDS-PAGE and dot blot.
Results
usp45-MSP-119 gene was amplified using specific primers and inserted into the multiple cloning sites in the expression vector pNZ8148 with size 3,538 bp as a recombinant vector. The protein of MSP-119 was successfully expressed in L. lactis with molecular weight of 10.45 kDa. The dot blot was tested in 3 different comparisons between the host cells, non-induced cells, and induced cells with 10 ng/ml nisin. The results showed that 10 ng/ml nisin gave a positive reaction as detected by dot blot assay.
Conclusion
This study confirmed that the usp45-MSP-119 gene was successfully inserted into the multiple cloning sites of the pNZ8148 expression vector and the MSP-119 protein expressed in the NICE system of the L. lactis host cell.
Keywords
Lactococcus lactis, malaria, merozoite surface protein 1, nisin, usp45-MSP-119
Malaria is one of the most contagious diseases that are still of a public health problem in the world including Indonesia. Malaria occurs in 106 countries. About 3.3 billion people live in endemic areas of malaria, and nearly 80% of malaria cases occur in the eastern part of Indonesia.1 The prevalence of malaria reflected as annual malaria incidence (AMI) decreased in the period 2000– 2004 from 31.09 to 21.2 per 1.000 population. In 2008, it fell to 18.82, and in 2010 it dropped to 12.27 per 1.000 population. Meanwhile, the prevalence of malaria annual parasite incidence (API) in 2000 was 0.81, and in 2004 fell to 0.15 per 1,000 population. In 2006, it increased to 0.19 then in the year 2007–2008 fell into 0.16 per 1,000 population.2
While currently artemisinin has been shown to be effective for malaria treatment, some degree of resistance has been reported to occur worldwide.3Therefore, a malaria vaccine is likely to be used as a potential tool for malaria control. Merozoite surface protein 1 (MSP-1) is a potential candidate of malaria vaccine because the monoclonal antibodies raised against this protein show its capability to block erythrocyte invasion in vitro.4 The MSP-1 is a major protein used by the Plasmodium during red blood cells invasion.5 Merozoite is the smallest cell measuring 1–2 μm in the life cycle of Plasmodium.6 The MSP-1 contains 2 epidermal growth factors (EGF-like domain) which are important parts during erythrocyte invasion.7,8 MSP-119 gene sequence located on block 17 is highly conserved. Therefore, it is used for the development of malaria vaccine.9
Lactococcus lactis has a generally recognized as safe (GRAS) host cell status and is often used in expressing recombinant proteins. L. lactis has been characterized as a good microorganism for the industry and capable of secreting recombinant protein into its growth media, so that the resulting product is free of endotoxin. The construction and expression of the MSP-119 gene in L. lactis has been prepared before.7 In the study, MSP-119 native gene construct was made by the fusion of SlpA peptide signals and intracellular protein expression. However, the level of MSP-119 protein expression is low. Estimated protein MSP-119 produced are only a few ng per 107 bacterial cells.
In addition, protein isolation is done through cell lysis. Since proteins are still at intracellular level, protein extraction process becomes more difficult. The fusion of the SlpA peptide signal should be able to bring the MSP-119 protein out through the cell membrane, so it can be secreted to the extracellular. In this study, the construction and expression of MSP-119 gene that has undergone codon optimization fused with usp45 signal peptide has been transformed into L. lactis. The usp45 signal peptide allows the MSP-119 protein to be secreted into the extracellular compartment. The usp45 signal peptide is an N-terminal signal peptide capable of optimizing the translocation of precursor proteins from within cells to the outside of the L. lactis cell wall.10 The codon optimization is performed to remove the biased codons that can inhibit the production of recombinant proteins in the prokaryotic system. It is a strategy to improve the production efficiency of recombinant proteins in the prokaryotic system. The purpose of this study was to construct the expression vectors for the MSP-119 gene and to express MSP-119 recombinant protein L. lactis as a candidate for malaria vaccine.10
METHODS
Genes, plasmids, bacteria, growth media, and primers used in this study
The material used in this study is the synthetic gene usp45-MSP-119 (GeneBank: AF165928) which is the result of codon optimization and has been ligated into the plasmid pMA-T. The growth media are Luria-Bertani (LB) and M17B bacteria. Plasmid used is pMA-T (cloning vector), and pNZ8148 is used as expression vector. Several vector and bacterial strains can be seen in Table 1, as well as several the primer pairs used in this study: forward ProNisA primer with TTCCCTCGAGGGATCTATGTC sequence (restriction site Nco1) and terminator reverse pNZ8148 primer with TGCTTTATCAACTGCTGC sequence (restriction site Xba1). E. coli was grown in Luria-Bertani (LB) medium (10 g/l tryptone [Oxoid, England], 10 g/l sodium chloride [Merck, Denmark], 5 g/l yeast extract [Oxoid, England]) supplemented with 100 μg/ml ampicillin (Bio Basic, Canada) at 37°C with agitation. Monoclonal antibody used for dot blot hybridization assay was Plasmodium falciparum MSP1 antibody (PEM-1) from Thermo Fisher. MSP-1 is highly conserved among broad Plasmodium species. Thus, this antibody could also detect MSP-1 from P. yoelii.
Table 1. Bacterial strains and vector used in this study

Construction of expression vector
The construction of the expression vector began by isolating the pMA-T plasmid. The MSP-119 gene was obtained by digesting the fragment of interest using restriction enzymes on the Nco1 and Xba1 sites, followed by gel purification. Pure DNA fragments of MSP-119 were ligated into pNZ8148 expression plasmid using T4 DNA ligase enzyme prior to introduction into E. coli MC1061. The transformation to E. coli MC1061 was performed by the heat shock method.11
Furthermore, pNZ8148-usp45-MSP-119 plasmid in E. coli MC1061 was isolated and subsequently transformed into L. lactis. Competent cells were grown in 0.2% L. lactis in 5 ml M17 broth + 0.5% glucose overnight at 30°C. Subsequently, the culture was incubated in 25 ml M17 broth containing 0.5% Glucose, 0.5 M sucrose, and 2.5% glycin overnight at 30°C. The culture was then put into 75 ml of M17 broth containing 0.5% glucose, 2.5% glycine, and 0.5 M sucrose and grown until the culture reached OD600 ~ 0.2–0.3 prior to harvesting the cell using centrifuge. Cells were resuspended with 0.5 M sucrose +10% glycerol. The transformation process was carried out using Gene Pulser BIORAD electroporation at 2,000 V, 25 μF, 260 Ω. The success of the transformation process of the pNZ8148-usp45-MSP-119 construct into L. lactis was confirmed by amplification of the bacteria colony by polymerase chain reaction (PCR), plasmid isolation followed by restriction enzyme digestion, and sequencing of inserts or plasmid construct.
pNZ8148-usp45-MSP-119 plasmid in L. lactis was grown further to increase the expression of the recombinant protein.12 L. lactis carrying recombinant plasmid was grown in 5 ml of M17 medium that contained 0.5% glucose. A total of 1.5 ml of L. lactis culture was subjected by centrifugation at 10.000x g for 2 min. The pellet was resuspended with solution I containing 10 mM EDTA (pH 8.0), 25 mM Tris/HCl, 50 mM glucose, and 20 mg/ml lysozyme (Sigma) was added and subsequently incubated for 15 min at 37°C. The mixture was added with 300 μl fresh solution II containing 0.2 M NaOH, 2% SDS and incubated for 3 min at room temperature. As much as 170 μl solution III containing 1.2 M Tris/HCl (pH 7.0) and 2 M NaCl were added to the mixture. The mixture was subsequently homogenized with 500 μl phenol. The upper layer phase was mixed with 600 μl PCI (Phenol Chloroform Isoamyl alcohol) then it was centrifuged. Moreover, 600 μl of isopropanol was added and then subjected to incubation on ice for 15 min. The mixtures were subjected to centrifugation and washed with 70% ethanol. The pellet was dried and dissolved in 20 μl ddH2O and 5 μl of 1 mg/ml RNase. The results of plasmid isolation were then subjected to Nco1 and Xba1 restriction enzymes, PCR of the colonies, and DNA sequencing to confirm the inserts sites. The schematic construction representation of the cloning vector pMAT and expression vector pNZ8148 which was expressed as a fusion protein pNZ8148- usp45-MSP-119 can be seen in Figure 1.

Figure 1. Schematic representation of the pNZ8148-usp45-MSP-119 expression construct containing the pMAT cloning vector and pNZ8148 expression vector
Expression and purification of recombinant protein MSP-119
Expression of MSP-119 as heterologous protein inside L. lactis was conducted with nisin inducible controlled expression (NICE) system. Protein expression was performed by the following methods. The MSP-119 expression in L. lactis was started by inoculating 10% fresh culture into 100 ml M17 broth containing 10 μg/ ml chloramphenicol and 0.5% glucose without agitation at 30°C for 10 hours to reach OD600 ~ 0.5. Furthermore, induction was performed using 10 ng/ml nisin and incubated for 5 hours at 30°C. The cells were subjected to centrifugation at 10.000 rpm at 4°C for 30 min. The recombinant protein was subjected to precipitation with 45% ammonium sulfate 27.7 g and subsequently incubated at 4°C for 10 hours. The cells were subjected to centrifugation to form the pellet for further elution step with 50 mM Tris HCl pH 7.4. The pellet suspension was purified by stationary phase adsorption chromatography Sephadex-G50. A total of 1 mL of pellet suspension was incorporated into chromatographic column, and then eluted with 50 mM Tris HCl pH 7.4.13,14
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis
The expressed proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis SDS-PAGE analysis using 10% separation gel and 4% retaining gel to examine the molecular weight of MSP-119. The recombinant protein was mixed with loading dye buffer (containing 50 mM Tris HCl pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoetanol, 12.5 M EDTA, 0.02% bromophenol blue in distilled water) and was subjected to heating at 95°C for 10 min prior to SDS-PAGE gel electrophoresis at 100 V and 20 mA. The gel was stained with silver staining kit (Fermentas Silver Stain Kit).
Determination of total protein concentration
The total protein concentration was quantified with bicinchoninic acid (BCA) kit using standard bovine serum albumine (BSA) curve. The standard concentrations of BSA used were 25 μg/ml, 125 μg/ml, 250 μg/ml, 500 μg/ml, 750 μg/ml, 1,000 μg/ml, 1,500 μg/ml, and 2,000 μg/ml. Working reaction was made by mixing reagents A and B with a ratio of 50:1. The sample and working reaction were added in a microplate 96-well by sample:working reaction ratio of 1:20. Microplate was incubated at 37°C for 30 minutes, and then the reaction result was read at 540 nm wavelength using enzyme-linked immunosorbent assay (ELISA) reader (Thermo Scientific Multiskan EX Microplate Reader).
Dot blot hybridization
Ten μl of protein samples were spotted on top surface of the membrane prior to blocking with 10% skim milk in 1x tris-buffered saline (TBS) for 1 hour. The membrane was washed 3 times, for 15 min, 5 min, and 5 min with 0.1% TBS-Tween. Furthermore, the membrane was incubated with monoclonal MSP1 antibody for 1 hour, and washed again 3 times with 0.1% TBS-Tween. The IgG mouse alkaline phosphatase conjugates as a secondary antibody was used to incubate the membrane for 1 hour prior followed by the washing step. Staining was performed using nitro blue tetrazolium-5-bromo-4-chloro-3-indolylphosphate NBT-BCIP substrate solution. The membrane was immersed in BCIP/NBT solution (500 μl NBT solution, 500 μl BCIP solution, dan 4 ml distilled water) for 1-5 min at room temperature until the color was developed. When the dot blots were detected, the incubation process was stopped by soaking the membrane in distilled water.15,16
RESULTS
The gene used in this study was the synthetic gene usp45-MSP-119 that has been ligated into the pMAT plasmid. The first stage of making the expression construct was the isolation of pMATusp45- MSP-119 plasmid with Nco1 and Xba1 restriction enzymes to release the gene/insert from the plasmid. The usp45-MSP-119 gene was then separated from the plasmid using restriction analysis as shown on agarose gel (Figure 2).

Figure 2. Restriction analysis of pMAT-usp45-MSP-119 recombinant plasmid M: 1kb DNA ladder marker, lane 1: digested pMAT plasmid of 2,757 bp, lane 2: undigested usp45- MSP-119 of 371 bp
Figure 3 describes the next stage was the ligation of the usp45-MSP-119 gene into the pNZ8148 expression vector. The 371 bp usp45-MSP-119 gene was successfully inserted into the multiple cloning site (MCS) of the pNZ8148 expression vector, forming a 3,538 bp recombinant plasmid. The recombinant vector was confirmed by Nco1 and Xba1 restriction enzymes. Ligation of the usp45-MSP-119 gene and the pNZ8148 vector was successfully transformed into E. coli MC1061.
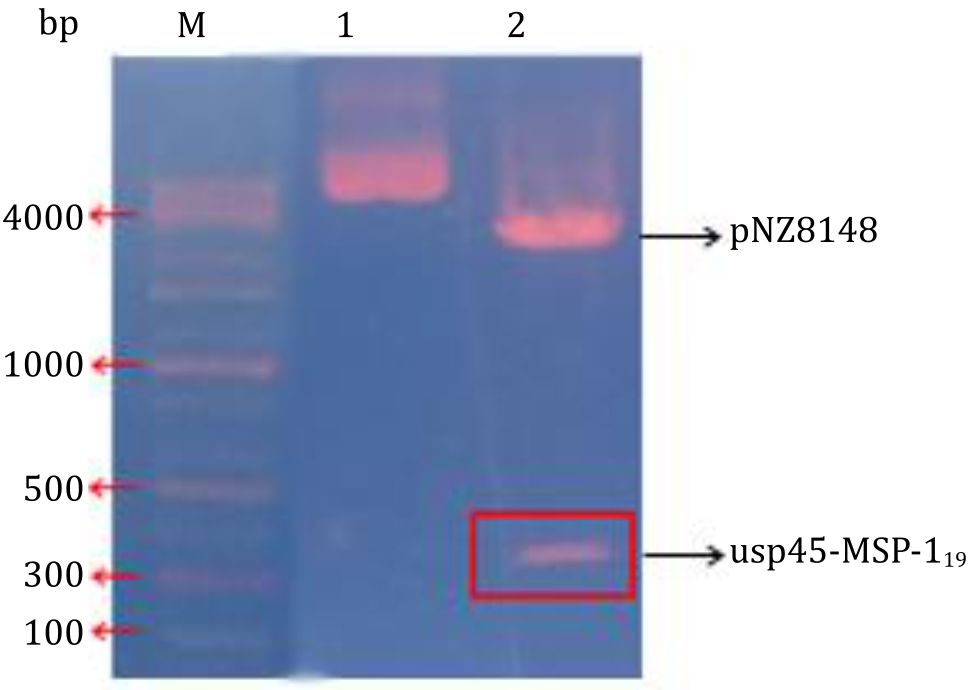
Figure 3. Confirmation of pNZ8148-usp45-MSP-119 recombinant protein by restriction enzyme cutting. Lane 1: undigested recombinant plasmid of pNZ8148 containing usp45- MSP-119 gene. Lane 2: pNZ8148- usp45-MSP-119 recombinant protein cut with Nco1 and Xba1 enzymes
The pNZ8148- usp45-MSP-119 recombinant vector was validated by DNA sequencing to determine whether or not mutations of the inserted MSP- 119 gene were present. The recombinant plasmid containing pNZ8148-usp45-MSP-119 was 3538 bp in size, encoding 124 amino acids, with a starting point of transcription from 47 nucleotides prior to the usp45-MSP-119 gene and ended at 559 nucleotides after the usp45-MSP-119 gene (Figure 4). The usp45-MSP-119 gene was cloned on the Nco1 site, causing the addition of 2 amino acids (6 nucleotides) during translation.
In this study showed the different concentration of total protein. The highest concentration of total protein was 0.936±0.04 mg/ml for cells that were induced with 10 ng/ml nisin. However, this was not significantly different with the concentration of total non-induced protein (0.549±0.03 mg/ml). The lowest concentration was the vector alone (0.188±0.04 mg/ml).
MSP-119 molecular weight was 10.45 kDa (Figure 5A) suggesting that the protein was expressed and secreted out of the cell due to addition of the usp45 signal peptide that was able to recognize the signal recognition particle (SRP). The expression of MSP-119 protein was shown by dot blot analysis tested by the MSP-119 monoclonal antibody as shown by the purple spot in the membrane (Figure 5B).
DISCUSSION
The expression vector construction in this study was initiated by the ligation of the usp45- MSP-119 synthetic gene into the pMAT plasmid. The inserts were verified using Nco1 and Xba1 restriction enzymes to separate plasmids and genes. The ligation of the usp45-MSP-119 gene into the pNZ8148 expression vector was done. The 371 bp usp45-MSP-119 gene was successfully inserted into the multiple cloning sites (MCS) in the pNZ8148 expression vector. Thus, it formed a recombinant protein of 3,538 bp.
The recombinant vector pNZ8148-usp45- MSP-119 was introduced into L. lactis NZ3900 using electroporation technique. This technique was used to insert the foreign DNA into a host of gram-positive bacteria that had the characteristic of a thick cell membrane structure.17 This electroporation technique applied high electrical voltage passed in a short time. Thus, it formed a pore on the cell membrane that could be inserted with foreign DNA. Selection of transformants using chlorampenicol antibiotics was possible because the vector harbors, the chloramphenicolresistant genes, was used as selection marker. The addition of sucrose and glycine can help the transformation process to become more efficient.18
The ligation of usp45-MSP-119 gene into pNZ8148 vector which was successfully introduced into E. coli host MC1061 was drawn from research by Wu et al.12 They introduced the vector into E. coli first and then to the L. lactis, so that the efficiency of the transformation to L. lactis was high. The pNZ8148 expression vectors are known to not have a high vector replication level, but it is balanced by their two starting points of replication, repA and repC, which are easily recognized by E. coli and L. lactis, respectively.19
Referring to Wu et al,12 the gene inserted into the expression vector was first adapted to the addition of two nucleotide bases of CC after translational starting site fused with Nco1 restriction sites (CCATGG). This is because the Nco1 site has already contained the ATG as the starting point of translation leading to mistranslation start site at the Nco1 site. At the usp45-MSP-119 signal peptide gene sequence in the pNZ8148 vector (Figure 4), there was NisA -35, -10, +1 as the transcription beginning of the ribosome-binding region/RBS (Shine-Dalgarno), TAA as the stop codon and it was terminated by a terminator as the ending point of transcription.20

Figure 4. Sequencing analysis pNZ8148- usp45-MSP-119 protein recombinant. NisA -35, -10, 1: are the starting points of transcription, RBS is the ribosome binding region (Shine-Dalgarno) and terminator is the end point of transcription. The green color is the restriction site (Nco1/Xba1), the blue color is Usp45 signal peptide, the red color is merozoite surface protein 1 (MSP-119), and the three asterisk is the stop codon
The usp45-MSP-119 gene was expressed in the nisin inducible expression (NICE) system. The expression of the MSP-119 recombinant protein was performed by the induction of 10 ng/mL nisin. Giving nisin as an inducer from the outside the cells will stimulate the start of transcription at the nisA promoter through nisK and nisR regulon integrated in chromosomal DNA of L. lactis.21 Possible expression level is controlled by the concentration of nisin given from the outside. The results of this response will form a transcription factor that activates the promoter nisA, so that the transcription process by RNA polymerase can be functioning.22
The presence of target protein bands in noninduced samples is expected due to basal activity causing the usp45-MSP-119 gene to be expressed even in the absence of nisin induction. This has been demonstrated in the Mohseni et al21 study that NICE expression systems often exhibit basal expression activity (in the absence of induction) when expressed in Lactobacillus. In addition to adjusting the dose of nisin, several other ways can be used to increase the gene expression level, such as by substituting an inducible promoter to a constitutive promoter. In accordance to the results of Lages et al,23 the administration of 10 ng/ml nisin induction in L. lactis increased gene expression, resulting in more protein than the non-induced. The concentrations of nisin has compared in the range between 0.5 and 5 ng/ml to induce the expression genes of antibacterial in L. lactis, and obtained the result that the highest of antibacterial was produced by L. lactis induced with nisin at 5 ng/ml by Mustopa et al.24 Moreover, codon optimization can also be performed. Nisin concentration required for induction is very low, between 0.5–10 ng/ml, while the high concentration of nisin of more than 10 ng/ml used will have negative effect on the host because it can suppress the growth of L. lactis.25
As stated in the results section, usp45 signal peptide was able to recognize signal recognition particle (SRP) indicated by the protein being successfully secreted outside the cell to the growing medium. The SRP will bring the prepeptide to the SRP receptor present in the membrane, which then will be brought to the protein translocation channel (PTC). The signal peptide opened the PTC, then the mature peptide was removed whereas the usp45 signal peptide would be cut by the peptidase.
Detection of MSP-119 protein was also seen from the dot blot hybridization test which is an immunological test whose function is similar to the western blot to detect the specificity of the reaction between the antigen and the antibody. In the samples induced with 10 ng/ml nisin, the positive results of the dot blot test were observed (Figure 5B). The results showed specific reactions between MSP-119 proteins with MSP1 monoclonal antibodies, seen from color developed on the membrane. Characterization of immunogenicity of primary antibodies tested by dot blot assays showed the result of 10.45 kDa MSP-119 antigen was reacted with anti-MSP1 specific antibodies.

Figure 5. Expression and purification of the MSP-119 fusion protein. A) SDS-PAGE of target protein expression and purification under different comparisons of induction, non-induced, and the lysed host cell. Molecular weight marker has shown in the left lane; lane 1: induced with 10 ng/ml nisin; lane 2: non-induced; lane 3: Lactococcus lactis; B) Dot blot analysis. Boxes 1, 2 and 3 are MSP-119 dot blots induced with 10 ng/ml nisin; Boxes 4 and 6 are MSP-119 dot blots non-induced; Box 5 is the dot blot of Lactococcus lactis
In conclusion, this study showed that MSP-119 protein fused with usp45 as signal peptide has been successfully constructed and expressed inside the GRAS organism in L. lactis. This heterologous expression of MSP-119 protein by GRAS host cell can be expected as the candidate for mucosal malaria vaccine which could induce both mucosal and systemic responses. Although initial test of expressed MSP-119 with monoclonal antibody in vitro has showed a positive result, in vivo investigation related to the efficacy of induced antibody should be determined.
Conflicts of Interest
The authors affirm no conflict of interest in this study
Acknowledgment
This study was supported by Research Centre for Biotechnology, Indonesian Institute of Science (LIPI), Bogor, Indonesia. The authors would like to thank Dr. Apon Zaenal Mustopa for all his supports.
REFERENCES
- who.int [Internet]. World Health Organization Malaria Fact Sheet 2011. Geneva: WHO Media Centre. [updated 2011, cited 2015 May]. Available from: www.who. int/malaria/world_malaria_report_2011/WMR2011_ factsheet.pdf
- Kemenkes RI. Pusat Data Informasi Kesehatan: Profil Kesehatan Indonesia. Jakarta: Kementerian Kesehatan Republik Indonesia; 2011. Indonesia.
- Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial drug resistance: literature review and activities and findings of the ICEMR network. Am J Trop Med Hyg. 2015;93(3):57–68.
- Lazaraou M, Patino JAG, Jennings RM, Mclntosh RS, Shi J, Howell S, et al. Inhibition of erythrocyte invasion and Plasmodium falciparum merozoite surface protein 1 processing by human immunoglobulin G1 (IgG1) and IgG3 antibodies. Infect Immun. 2009; 77(12):5659–67.
- Baldwin MR, Li X, Hanada T, Lui S-C, Chisthi AT. Merozoite surface protein 1 recognition of host glycophorin A mediates malaria parasite invasion of red blood cells. Blood. 2015;125(17):2704–11.
- Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol. 2012; 198(6):961–71.
- Zhang ZH, Jiang PH, Li NJ, Shi M, Huang W. Oral vaccination of mice against rodent malaria with recombinant Lactococcus lactis expressing MSP-119. World J Gastroenterol. 2005;11(44):6975–80.
- Curd RD, Birdsall B, Kadekoppala M, Ogun SA, Kelly G, Holder AA. The structure of Plasmodium yoelli merozoite surface protein 119, antibody specificity and implications for malaria vaccine design. Open Biol. 2014;4:130091.
- Cruz-Gallardo I, Diaz-Moreno I, Diaz-Quintana A, Donaire A, Velazquez-Campoy A, Curd RD, et al. Antimalarial activity of cupredoxins: the interaction of Plasmodium merozoite surface protein 119 (MSP119) and rusticyanin. J Biol Chem. 2013; 288(9):20896-907.
- Ng DTW, Sarkar CA. Engineering signal peptides for enhanced protein secretion from Lactococcus lactis. Appl Environ Microbiol. 2013;79(1):347-56.
- Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th ed. New York (US): Cold Spring Harbor Laboratory Press; 2012.
- Duan K, Dunn NW, Kim WS. Rapid plasmid DNA isolation from Lactococcus lactis using overnight cultures. Biotechnol Tech. 1999;13:519–21.
- Wu C, Zhang J, Du G, Chen J. Heterologous expression of Lactobacillus casei RecO improved the multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 during salt stress. Bioresour Technol. 2013;143:238–41.
- Todorov SD, Ho P, Vaz-Velho M, Dicks LMT. Characterization of bacteriocins produced by two strains of Lactobacillus plantarum isolated from Beloura and Chouriço, traditional pork products from Portugal. Meat Sci. 2010;84(3):334–43.
- Pertiwi W, Sartono TR, Sumarno, Adi S. Sensitivitas dan spesifisitas metode dot blot menggunakan antigen outer membrane protein Klebsiella pneumoniae yang direspon secretory-immunoglobulin A sputum penderita terinfeksi Klebsiella pneumoniae. J Respir Indones. 2009;29(3):1–15.
- Widjiati, Pradipta AR, Nazar DS, Estoepangestie ATS. Uji spesifisitas dengan dot blotting terhadap epidermal growth factor (EGF) yang diisolasi dari oosit kumulus komplek sapi setelah dimaturasi secara in vitro. Vet Med. 2014;7(2):134–9. Indonesian.
- . Rattanachaikunsopon P, Phumkhachorn P. Glass bead transformation method for gram-positive bacteria. Braz J Microbiol. 2009;40(4):923–6.
- Heravi RM, Nasiraii R, Sankian M, Kermanshahi H, Varasteh AR. Optimization and comparison of two electrotransformation methods for Lactobacilli. Biotechnology 2012;11(1):50–4.
- De Ruyter PGGA, Kuipers OP, de Vos WM. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62(10):3662–7.
- Korkmaz G, Holm M, Wiens T, Sanyal S. Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance. J Biol Chem. 2014;289(44):30334–42.
- Mohseni AH, Razavilar V, Keyvani H, Razavi MR, Khavari-Nejad RA. Efficient production and optimization of E7 oncoprotein from Iranian human papillomavirus type 16 in Lactococcus lactis using nisin-controlled gene expression (NICE) system. Microb Pathog. 2017;110:554–60.
- Wang ZH, Wang YL, Zeng XY. Construction and expression of a heterologous protein in Lactococcus lactis by using the nisin-controlled gene expression system: the case of the PRRSV ORF6 gene. Genet Mol Res. 2014;13(1):1088–96.
- Lages AC, Mustopa AZ, Sukmarini L, Suharsono. Cloning and expression of plantaricin w produced by Lactobacillus plantarum U19 Isolate from “Tempoyak” Indonesian fermented food as immunity protein in Lactococcus lactis. Appl Biochem Biotechnol. 2015;177:909–22.
- Mustopa AZ, Murtiyaningsih H, Fatimah, Suharsono. Cloning and heterologous expression of extracellular Plantaricin F produced by Lactobacillus plantarum S34 isolated from “Bekasam” in Lactococcus lactis. Microbiol Indones. 2016;10(3):95–106.
- Zhang X-J, Feng S-Y, Li Z-T, Feng Y-M. Expression of Helicobacter pylori hspA gene in Lactococcus lactis NICE system and experimental study on its immunoreactivity. Gastroent Res Pract. 2015;2015:1–6.
Copyright @ 2017 Authors. This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original author and source are properly cited.
mji.ui.ac.id