
Section Abstract Introduction Methods Results Discussion Conflict of Interest Acknowledgment References
Clinical Research
Molecular survey on Merkel cell polyomavirus in patients with colorectal cancer
pISSN: 0853-1773 • eISSN: 2252-8083
https://doi.org/10.13181/mji.v27i4.2759 Med J Indones. 2018;27:229–36
Received: April 7, 2018
Accepted: September 24, 2018
Author affiliation:
1 Institute of Immunology and Infectious Diseases, Iran University of Medical Sciences, Tehran, IR Iran
2 Gastrointestinal and Liver Disease Research Center, Iran University of Medical Sciences, Tehran, IR Iran
3 Firoozgar Clinical Research Development Center (FCRDC), Iran University of Medical Sciences, Tehran, IR Iran
4 Department of Virology, Pasteur Institute of Iran, Tehran, IR Iran
5 Department of Virology, Iran University of Medical Sciences, Tehran, IR Iran
6 Iranian Biological Resource Center, Microorganism’s Bank, Tehran, IR Iran
Corresponding author:
Hossein Keyvani
E-mail: keyvanlab@yahoo.com
Background
Merkel cell polyomavirus (MCV) has been associated with Merkel cell carcinoma (MCC) in humans, and its role in other human cancers is under investigation. The aim of this study was to investigate MCV genome infection in patients with colorectal cancer (CRC).
Methods
This retrospective, case-control study used archived formalin-fixed, paraffin-embedded (FFPE) tissue samples from colorectal cancer patients (cases) and matched healthy subjects (controls) diagnosed by an expert pathologist from hospitals affiliated with Iran University of Medical Sciences, Tehran, Iran from 2011 to 2016. After DNA extraction with a QIAamp® DNA FFPE Tissue Kit, real-time polymerase chain reaction (PCR) was used for diagnosis. A positive control was produced by cloning with the Generay Biotechnology system. SPSS v.22 was used for analysis of demographic variables.
Results
There were 157 participants included in the study: 66 were cases and 91 were controls. Their mean ages (±SD) were 59.35±14.48 and 57.21±14.66, respectively. The proportion of males was 57.6% in the case group and 57.1% in the control group. None of the samples were positive for MCV expression by real-time PCR assay. Association was detected between males with CRC and tumor location in the rectum and between males with CRC and the mucinous tumor type.
Conclusion
None of the tissues from the CRC or non-cancerous control groups were positive for MCV genome infection, although a low viral load, the sample type, or the method of use should not be neglected. Further studies are recommended to obtain more comprehensive results.
Keywords
cancer, colorectal, Merkel cell polyomavirus, real-time PCR, viral infection
Cancer is a life-threatening disease in humans worldwide.1,2 Colorectal cancer (CRC) is the third most common cancer and includes 10% of all cases of cancer globally.1,2 Most CRC are caused by factors associated with lifestyle and age, whereas few occur due to genetic and hereditary disorders.2 Viral infections are responsible for approximately 10–15% of human cancers worldwide.3–5 Studies have shown that viruses from the Polyomaviridae family are oncogenic in animal models and are capable of transforming cells in culture.6 BK virus, John Cunningham (JC) virus, Simian virus 40 (SV40), and the recently known Merkel cell polyomaviruses (MCPyV or MCV) have nonenveloped virions of 45–50 nm diameter.7-8 They have a circular double-stranded DNA genome with an approximate size of 5 kilobase pairs (kbp).9 Generally, polyomavirus (PyV) lytic infection is responsible for their pathogenesis, but primary infection does not usually lead to an acute illness, and low levels of the virus establish a latent infection.10 A lack of cell culture models for PyV restrict us to the use of molecular diagnostics methods.11 The presence of PyVs in cancerous patients is associated with immunosuppression and a higher risk severe PyV disease; therefore, these patients should be monitored carefully and managed appropriately through their treatment course.10
Some polyomaviruses, including BK, JC, and SV40, have been investigated for their association with CRC.10,12 After some evidence of non-neuronal JC virus (JCV) complications in gastric and lung cancers came to light, Laghi et al described the potential risk of JCV in CRC and reported that 96% of tissues from CRC patients were positive for JCV genomic DNA.13,14 However, another study reported a rate JCV infection in CRC tissue of about 18.2%, and yet another study reported an infection rate of 100% in anal cancer tissue.10,15 However, according to a systematic review by Chen et al,5 some studies reported the absence of BK and JC genomes in CRC tissue.
Recently, MCV has gained attention due to its association with rare human cancers.7 MCV infection is common in the human population and is detected in various locations, such as skin of neuroendocrine cell origin.7,16 Given the carcinogenicity of MCV, the relationship between infection by this virus and various cancers, including skin tumors, non-Merkel skin tumors in neuroblastoma, central nervous system tumors, lung cancer and mesothelioma, has been investigated.11,17,18 However, in some high-grade neuroendocrine tumors of different body sites, there were no positive results for MCV DNA.18 Integration of the MCV genome in Merkel cell carcinoma (MCC) could stimulate cancer progression.18,19 Mutation or deletion of nucleotide sequences in the early genomic region could produce a truncated T antigen, impaired replication of the viral genome, and cell transformation.18
Some researchers have identified the MCV genome in CRC specimens, but currently there is not enough information about the role of MCV in CRC.5,18 The rationale for investigation MCV in CRC is the probable similar epidemiological pattern and association of some PyVs in CRC.20 Studies investigating the possible role of MCV in CRC are rare, and the majority of explorations were performed comparing cancer tissues with pre-cancerous lesions and did not include control subjects.18,20 In addition, there are no data on the prevalence of MCV in the Iranian gastrointestinal cancer patient population. Accordingly, this study sought to investigate the presence of the MCV genome in CRC compared with pre-cancerous healthy tissue lesions as a control group.
METHODS
Study population
placed in −80°C for 5 min, and this cycle was continued In this retrospective, case-control study, patients diagnosed by expert pathologists from hospitals affiliated with Iran University of Medical Sciences, Tehran, Iran from 2011 to 2016 were included. Consent was obtained from each patient. Ethical approval was obtained from the ethics committee of Iran University of Medical Sciences, Tehran, Iran by the code: IR.IUMS. FMD.REC 1394.26295. Archived formalin-fixed, paraffin-embedded (FFPE) blocks were collected from patients with CRC (66 cases) and matched healthy subjects (91 cases) as controls. All subjects underwent endoscopy and colonoscopy with the use of a Fujinon endoscope (Fujinon, Japan). Patient data were collected from the medical record repository. Included CRC patients were sporadic, non-familial cancerous cases and gave written consent. The healthy control group included participants who underwent total colonoscopy, matched by sex and age with the case group, and had no malignant lesions. Slides stained with hematoxylin and eosin (H&E) were reviewed by an experienced pathologist, and 20-micron-thick slices were used for all samples.1
DNA extraction
The QIAamp® DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) was used for genomic DNA extraction from CRC patients and healthy pre-cancerous lesions patients.21,22 Purified DNA was quantified by using a NanoDrop ND-1000® (Thermo Fisher Scientific Inc., Waltham, MA, USA) spectrophotometry. DNA was kept at −20˚C before use.
Real-time polymerase chain reaction (PCR)
A real-time PCR assay was optimized for evaluation of the T-Large Ag (LT) of MCV by the SYBR Green method. Specific forward (5’-TCTTCCTCTGGGTATGGGTC-3’) and reverse (5’-ATTGGGTGTGCTGGATTCTC-3’) primers for LT Ag from previous authors works were used (17, 23). The Rotor-Gene-Q 6000 thermocycler (Corbett, Australia) was used for real-time PCR assay optimization. The melting curve was analyzed to investigate the reaction specificity (PCR product differentiation from non-specific amplification). The components of the total reaction mixture were 2× Amplicon III mix (Odense M, Denmark) 7.5 μl, each forward and reverse primer to a 0.5 μM concentration, each sample or control to a 0.2–0.5 μM concentration, and distilled water to obtain a total volume of 15 μl. To increase the accuracy of the analysis, a duplicate test was performed for each test sample. The cycling program used consisted of an initial denaturation for 5 min at 95˚C and 40 cycles of 30s at 95˚C and 30s at 57˚C; acquisition at annealing extension was performed. Fivesecond Intervals from 50˚C to 99˚C were used for melting curve analysis. A positive control was obtained from the work of previous authors, and its quality was confirmed by cloning and realtime PCR methods.17–23 The full-length MCV LT Ag target of the forward and reverse primers was cloned and analyzed on the basis of previously published protocols.18,24
Statistical analysis
All data were entered into Statistical Product and Service Solutions (SPSS) software version 22. Chi-square analysis or Fisher’s exact test were used to evaluate the data with regard to categorical and qualitative variables such as lymph node involvement or tumor stage. A p-value ≤0.05 was considered to indicate statistical significance.
RESULTS
Study population
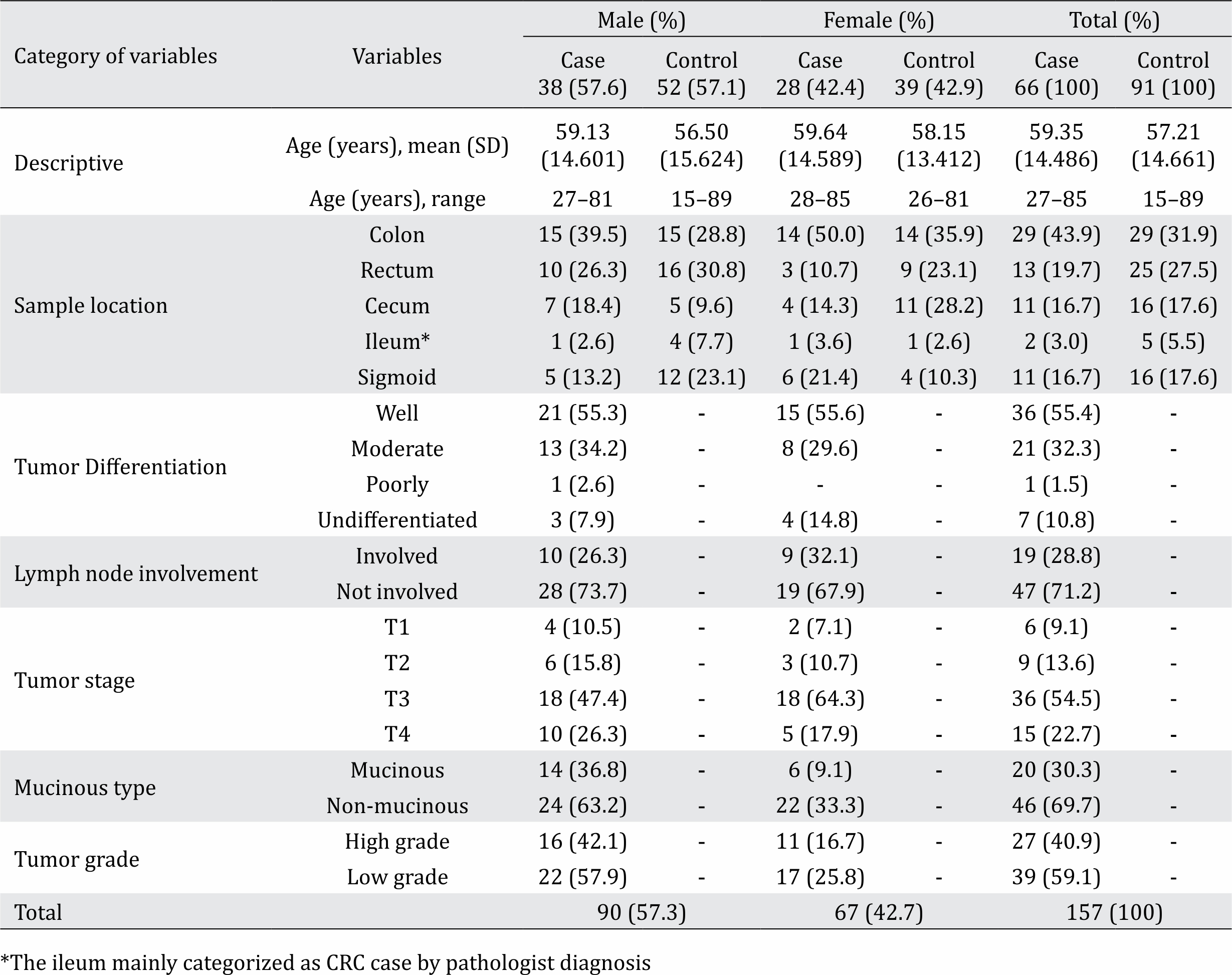
A total of 157 specimens including 66 CRC cases and 91 healthy controls participated in this study. Of the 61 cases, the mean age (±SD) was 59.35±14.49 and 57.6% were male. Of the 91 healthy participants, the mean age (±SD) was 57.21±14.66 and 57.1% were male. Demographic and pathologic characteristics of the case and control groups are shown in Table 1.
Table 1. Demographic and pathologic characteristics of our study population (n=157)
Real-time PCR assay
All 157 samples were analyzed by realtime PCR assay to determine the prevalence of MCV by using specific LT Ag primers. Although the positive and negative controls produced a reasonable amplification curve (not shown), none of the specimens produced a positive signal. A melting curve diagram showed the specificity of the primers and conditions for MCV detection. Confirmation of the positive control by specific primers that were used in the current study was carried out by PCR, and the result is shown in Figure 1. Plasmid sequencing results for the positive control are shown in Figure 2.
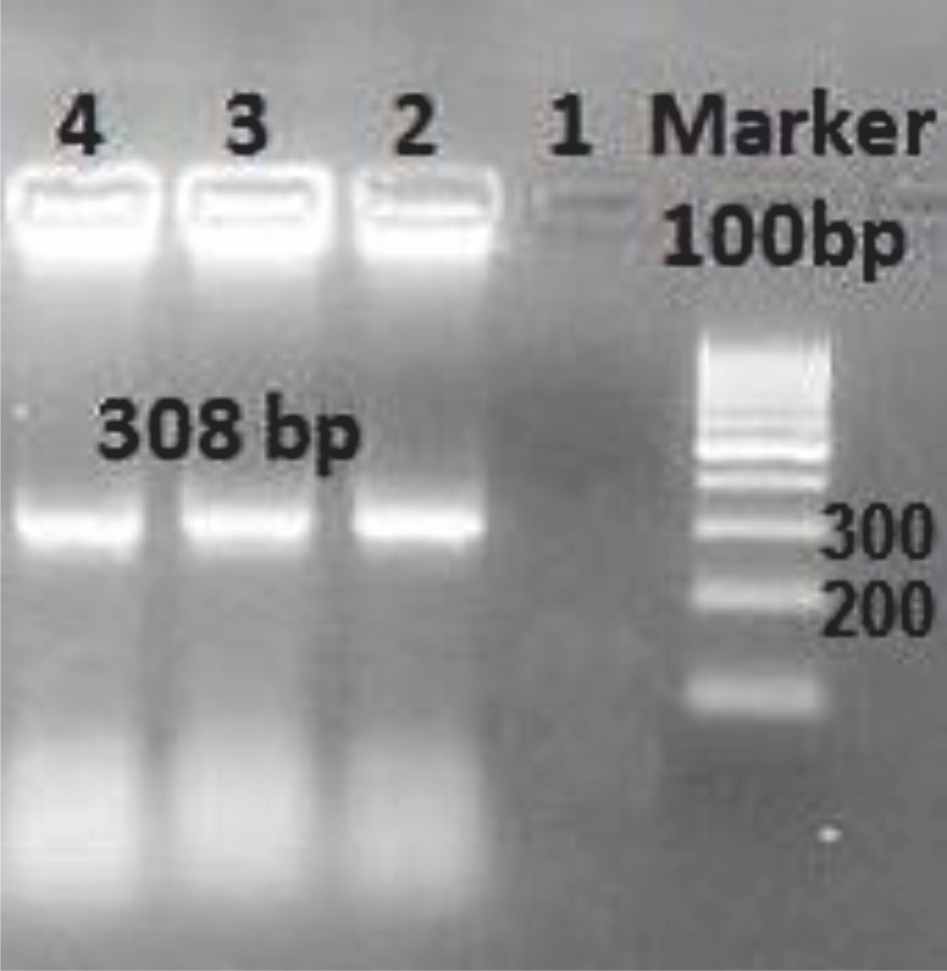
Figure 1. PCR confirmation of MCV LT Ag cloned into the pTZ57R/T plasmid as a positive control with the specific primers used in the present study. 1-3: triplicate plasmid clones that were used as positive controls; 3: non-template negative control
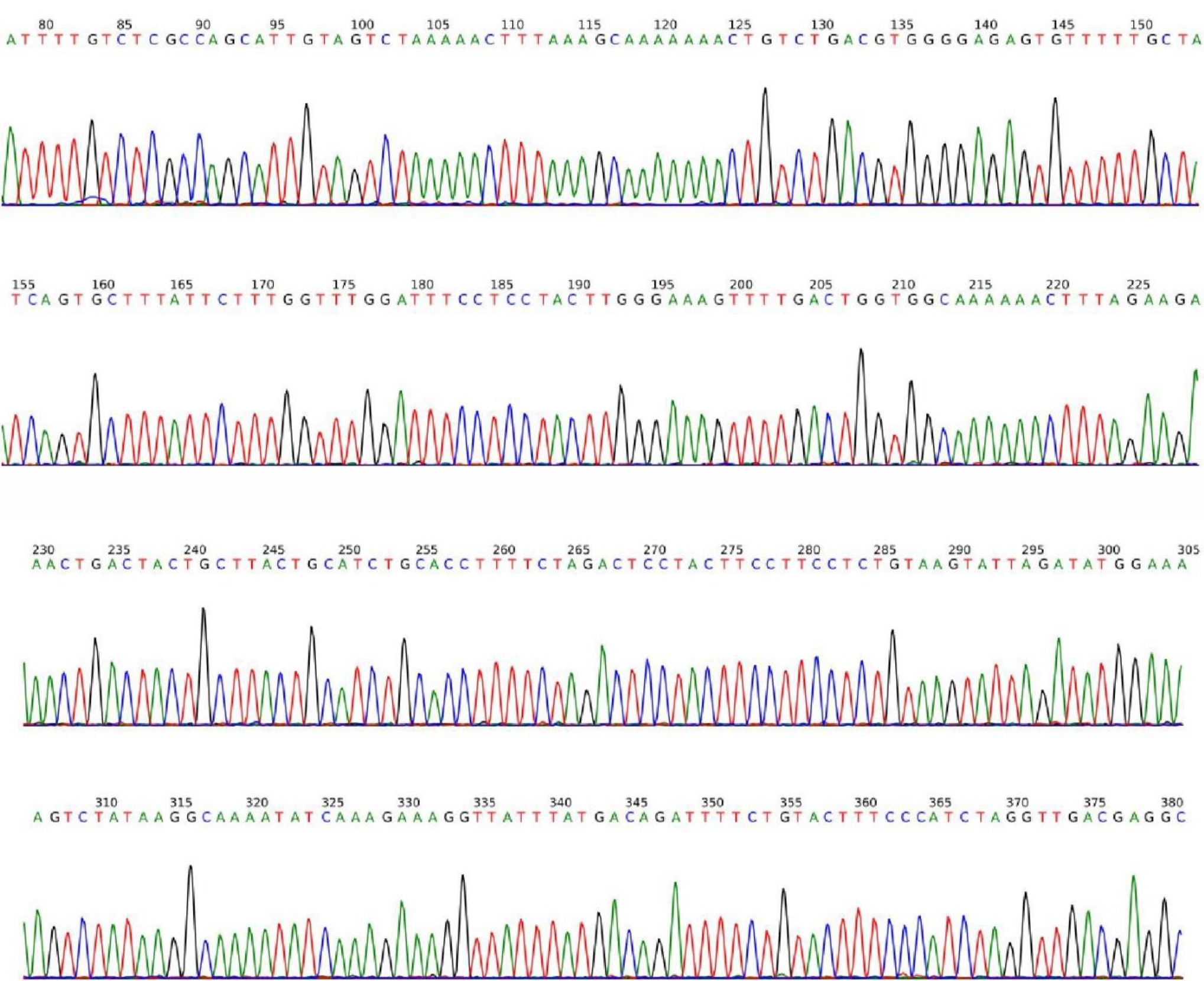
Figure 2. Plasmid sequence of MCV LT Ag cloned into pTZ57R/T, which was used as positive control
Statistics
Statistical analyses of different demographic variables by pathologic data showed no significant results in the comparison of CRC outcome with gender, although CRC was more often seen in males. Rectum location and mucinous type of CRC in males were statistically significant (p≤0.05).
DISCUSSION
The relationship between polyomavirus infections and gastrointestinal malignancies is under investigation.18 In recent studies, the JCV and MCV genomes have been identified in sewage and environmental samples, which indicates the possibility of transmission of these viruses by the oral-fecal route.24 The targets of MCV could be epithelial, digestive, nervous system, and sometimes kidney tissues.8 Rapid diagnosis of MCV in colonic adenoma is important for choosing appropriate intensive treatment strategies for these patients.11 A molecular-based therapy could improve clinical outcomes and reduce disease severity. Based on our findings by the present study, it is not applicable to Iranian CRC population, although our limited population or potential inhibitors from human tissue could have impacted on the infection detection frequency.
Polyomavirus-associated CRC might be transferred by meat as hypothesized by zur Hausen.25 Beef has been shown to contain different human and animal polyomaviruses, such as MCPyV, human polyomavirus 7 (HPyV7), and raccoon polyomavirus (RacPyV).25 MCV, like some other polyomaviruses, secretes tumor antigen (T-Ag) oncoproteins that could readily induce tumor formation.17 One possible mechanism of MCV oncogenesis is based on the LT and sT antigens that downregulate Toll-like receptor 9 (TLR9), an important receptor for the innate immune system. This downregulation could suppress the CCAAT-enhancer binding protein β (C/EBPβ) transcription factor and cause further defects in immune responses and consequences for cell proliferation.26 Another proposed mechanism is sT antigen downregulation of NF-kB-mediated transcription and further triggering of carcinogenesis.26 Moreover, there are some functional domains in the sT Ag that present a DnaJ domain and contain zinc (Zn) binding sites. The PPAR1/NEMO binding site and a large T stabilization domain in MCV have roles in oncogenic processes.27 Although some researchers have identified the genome of MCV polyomavirus in CRC patients, currently, there is not enough information about the role of MCV polyomavirus in CRC.5,7 In addition, there is no information on the prevalence of MCV in samples of gastrointestinal cancers in the Iranian population. Therefore, this study aimed to investigate the presence of the MCV genome in CRC patients. Our studied samples included 66 subjects with CRC and 91 healthy individuals who underwent total colonoscopy and had noncancerous masses. After DNA extraction, real-time PCR was conducted by using LT Ag region-specific primers. According to the results of the present study, none of the samples were positive based on the confirmed positive and negative controls when specific T antigen primers were used. It is likely that that the level of MCV infection in our community is low, especially in people with CRC complications, although there have been some rare reports of MCV infection in different malignancies in Iran.9,7,23,28–30 Also, MCV genome positivity was reported in 40%–100% of cutaneous swabs from clinically healthy subjects, and MCV was detected in respiratory secretions, on oral and anogenital mucosa, and in the digestive tract.31 Our negative results may also have been caused by inhibitors from human tissue or the formalin fixation step performed in FFPE blocks.
Some reports showed viral capsid protein antibodies to MCV in healthy subjects, although the same is true about other known human polyomaviruses (BK, JC, KI, and WU viruses).31–33 In one such study, MCV seroprevalence was 0% in infants, 43% in children 2–5 years of age, and up to 80% in adults older than 50 years.34 Studies showed 87.1% MCV antibody prevalence in adulthood,35 increasing from 45% in children under 10 up to 81% in adults older than 60 years, and seroprevalence of 91% in MCC patients, compared with 68% in a control group.36 The current study did not evaluate anti-MCV antibody positivity, which is recommended in further studies to obtain more comprehensive results, although anti-MCV antibody prevalence might be omitted since it has no correlation with CRC nor with the pathogenesis of CRC.
Some studies in Iran detected MCV in different malignant and non-malignant populations. Sadeghi et al17 in one such Iranian study reported an MCV prevalence of 59% in patients with central nervous system (CNS) tumors, and they indicated the existence of a low MCV viral load. Another study in Iranian patients by Salehi-Vaziri et al9 detected 33% MCV positivity in cervical specimens and 12.5% in coinfection with HPV. They concluded that MCV co-infection with HPV could lead to progression of malignancy. Mohebbi et al28 in head and neck squamous cell carcinoma patients showed 16% prevalence of MCV infection. Although they found a significant association between an increased viral load and a higher cancer stage, they concluded that its association should be investigated further. Another study by Yahyapour et al29 on esophageal squamous cell carcinoma (ESCC) showed that MCV DNA was detected in 45% of ESCC cases and 35% of non-malignant controls. Based on the literature, studies in other malignancies have been reported frequently. Baez et al37 reported a case of MCV DNA in a penile cancer patient. A study by Katano et al19 on MCC patients showed MCV positivity in 55% and 6.1% in Kaposi’s sarcoma patients, but did not detect it in autopsy samples from immunodeficient patients. Our study showed no positive results for the presence of the MCV genome in the studied population; further studies are required to verify the low rates of MCV infection in Iranian CRC patients. Furthermore, using more precise methods, such as nested PCR, or using fresh samples can be helpful when the viral load is low.
In CRC patients, Campello et al18 designed a case-control study to investigate MCV in fresh tissue from 64 CRC patients and fresh biopsies from 80 matched relatives. They also examined 144 blood samples. After using PCR and sequencing, they reported that 6.3% of cases and 8.8% of controls were infected by MCV. Blood samples showed an overall rate of MCV infection of 12.5%. This result showed a low frequency of MCV in the colon, although they used fresh samples for analyses. The present study used FFPE samples, and we could not detect any MCV positive specimens. A low copy number of the MCV genome based on previous studies and using FFPE blocks could have affected our results. Also, due to the retrospective study design and not having any blood samples, we were not able to verify our results serologically. Our population had similar ages in the CRC group to those in the study by Campello et al,18 which showed that other confounding effects or different socioeconomic factors could produce negative results in the detection of MCV, although there was a detectable frequency of MCV in our country in different populations.9,17,28,29
This study did not detect MCV infection in cancerous and non-cancerous colorectal specimens, a negative result could have been due to the low prevalence rates of the virus in the community or CRC population and/or a low viral load in the studied samples. Confirmatory studies using a larger sample size and different specimen types are recommended for a more thorough understanding of the MCV infection prevalence in CRC.
Conflicts of Interest
The authors affirm no conflict of interest in this study.
Acknowledgment
We are thankful for kind assistance of Keyvan laboratory, Tehran, Iran, personnel. This study conducted as the grant project of 94- 02-128-26295 from Iran University of Medical Sciences.
REFERENCES
- Koochak A, Rakhshani N, Karbalaie Niya MH, Tameshkel FS, Sohrabi MR, Babaee MR, et al. Mutation analysis of KRAS and BRAF genes in metastatic colorectal cancer: a first large scale study from Iran. Asian Pac J Cancer Prev. 2016;17(2):603–8.
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RG, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–93.
- Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15(3):266–82.
- Chiocca EA, Rabkin SD. Oncolytic viruses and their application to cancer immunotherapy. Cancer Immunol Res. 2014;2(4):295–300.
- Chen H, Chen XZ, Waterboer T, Castro FA, Brenner H. Viral infections and colorectal cancer: a systematic review of epidemiological studies. Int J Cancer. 2015;137(1):12–24.
- Teklemariam AD, Geleta G, Tassew A. A review on gene involved in cancer development and oncogenic viruses. Afr J Basic Appl Sci. 2015;7(4):223–32.
- MacDonald M, You J. Merkel cell polyomavirus: A new DNA virus associated with human cancer. Infectious agents associated cancers: Epidemiology and Molecular Biology: Springer; 2017. p. 35–56.
- Scadden JR, Sharif A, Skordilis K, Borrows R. Polyoma virus nephropathy in kidney transplantation. World J Transplant. 2017;7(6):329–38.
- Salehi-Vaziri M, Sadeghi F, Alamsi-Hashiani A, Haeri H, Monavari SH, Keyvani H. Merkel cell polyomavirus and human papillomavirus infections in cervical disease in Iranian women. Arch Virol. 2015;160(5):1181–7.
- Loutfy SA, Moneer MM, Salem SE, El-Moniem Abada EA, El-Moniem Ahmed EA, Ibrahim LH, et al. Polyomavirus infections and its clinical relevance in cancer patients: a prospective study. J Infect Public Health. 2017;10(1):22–30.
- Arora R, Chang Y, Moore PS. MCV and Merkel cell carcinoma: a molecular success story. Curr Opin Virol. 2012;2(4):489–98.
- Ripple MJ, Parker Struckhoff A, Trillo-Tinoco J, Li L, Margolin DA, McGoey R, et al. Activation of c-Myc and Cyclin D1 by JCV T-Antigen and β-catenin in colon cancer. PLoS One. 2014;9(9):e106257.
- Ramamoorthy S, Devaraj B, Miyai K, Luo L, Liu YT, Boland CR, et al. John Cunningham virus T‐antigen expression in anal carcinoma. Cancer. 2011;117(11):2379–85.
- Laghi L, Randolph AE, Chauhan DP, Marra G, Major EO, Neel JV, et al. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci U S A. 1999;96(13):7484–9.
- Mou X, Chen L, Liu F, Lin J, Diao P, Wang H, et al. Prevalence of JC virus in Chinese patients with colorectal cancer. PLoS One. 2012;7(5):e35900.
- Kisseljov FL, Vinokurova SV, Kisseljova NP. Novel human DNA viruses and their putative associations with human diseases. Mol Biol (Mosk). 2016;50(4):551–66.
- Sadeghi F, Salehi-Vaziri M, Alizadeh A, Ghodsi SM, Bokharaei-Salim F, Fateh A, et al. Detection of Merkel cell polyomavirus large T-antigen sequences in human central nervous system tumors. J Med Virol. 2015;87(7):1241–7.
- Campello C, Comar M, D’Agaro P, Minicozzi A, Rodella L, Poli A. A molecular case–control study of the Merkel cell polyomavirus in colon cancer. J Med Virol. 2011;83(4):721–4.
- Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, et al. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi’s sarcoma. J Med Virol. 2009;81(11):1951–8.
- Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, et al. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. Int J Cancer. 2010;126(12):2991–6.
- Eftekhaar NS, Karbalaie Niya MH, Izadi F, Teaghinezhad-S S, Keyvani H. Human papillomavirus (HPV) genotype distribution in patients with recurrent respiratory papillomatosis (RRP) in Iran. Asian Pac J Cancer Prev. 2017;18(7):1973–6.
- Karbalaie Niya MH, Safarnezhad Tameshkel F, Panahi M, Bokharaei Salim F, Monavari SH, Keyvani H. Human papillomavirus investigation in head and neck squamous cell carcinoma: initial report from the low risk HPV types associations. Asian Pac J Cancer Prev. 2017;18(9):2573–9.
- Sadeghi F, Ghanadan A, Salehi-Vaziri M, Salim FB, Monavari SH, Keyvani H. Merkel cell polyomavirus infection in a patient with merkel cell carcinoma: a case report. Jundishapur J Microbiol. 2015;8(3):e17849.
- Hamza H, Hamza IA. Oncogenic papillomavirus and polyomavirus in urban sewage in Egypt. Sci Total Environ. 2018;610–611:1413–20.
- zur Hausen H. Red meat consumption and cancer: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int J Cancer. 2012;130(11):2475–83.
- Moens U, Rasheed K, Abdulsalam I, Sveinbjørnsson B. The role of Merkel cell polyomavirus and other human polyomaviruses in emerging hallmarks of cancer. Viruses. 2015;7(4):1871–901.
- Baez CF, Brandão Varella R, Villani S, Delbue S. Human polyomaviruses: the battle of large and small tumor antigens. Virology (Auckl). 2017;8:1–12.
- Mohebbi E, Noormohamadi Z, Sadeghi-Rad H, Sadeghi F, Yahyapour Y, Vaziri F, et al. Low viral load of Merkel cell polyomavirus in Iranian patients with head and neck squamous cell carcinoma: is it clinically important? J Med Virol. 2018;90(2):344–50.
- Yahyapour Y, Sadeghi F, Alizadeh A, Rajabnia R, Siadati S. Detection of Merkel cell polyomavirus and human papillomavirus in esophageal squamous cell carcinomas and non-cancerous esophageal samples in Northern Iran. Pathol Oncol Res. 2016;22(4):667–72.
- Reza MA, Reza MH, Mahdiyeh L, Mehdi F, Hamid ZN. Evaluation frequency of Merkel cell polyoma, epstein- Barr and mouse mammary tumor viruses in patients with breast cancer in Kerman, southeast of Iran. Asian Pac J Cancer Prev. 2015;16(16):7351–7.
- Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Current Oncol Rep. 2011;13(6):488–97.
- Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5(3):e1000363.
- Touzé A, Le Bidre E, Laude H, Fleury MJ, Cazal R, Arnold F, et al. High levels of antibodies against merkel cell polyomavirus identify a subset of patients with merkel cell carcinoma with better clinical outcome. J Clin Oncol. 2011;29(12):1612–9.
- Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, et al. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer. 2009;125(6):1250–6.
- Nicol JT, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, et al. Age-specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clin Vaccine Immunol. 2013;20(3):363–8.
- Viscidi RP, Rollison DE, Sondak VK, Silver B, Messina JL, Giuliano AR, et al. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin Vaccine Immunol. 2011;18(10):1737–43.
- Baez CF, da Rocha WM, Afonso LA, Carestiato FN, Guimarães MA, Cavalcanti SM, et al. First report of three major oncogenic viruses: human papillomavirus, epstein-barr virus and merkel cell polyomavirus in penile cancer. J Infect Dis Ther. 2015;3(5):233.
Copyright @ 2018 Authors. This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original author and source are properly cited.
mji.ui.ac.id